8CYU
 
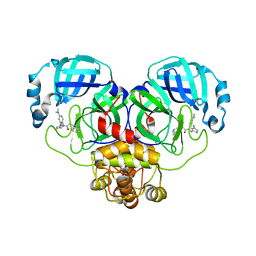 | | Crystal structure of SARS-CoV-2 Mpro with compound C5 | | Descriptor: | 3C-like proteinase, N-[(4-chlorothiophen-2-yl)methyl]-N-[4-(dimethylamino)phenyl]-2-(isoquinolin-4-yl)acetamide | | Authors: | Worrall, L.J, Lee, J, Strynadka, N.C.J. | | Deposit date: | 2022-05-24 | | Release date: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A novel class of broad-spectrum active-site-directed 3C-like protease inhibitors with nanomolar antiviral activity against highly immune-evasive SARS-CoV-2 Omicron subvariants.
Emerg Microbes Infect, 12, 2023
|
|
8CZ4
 
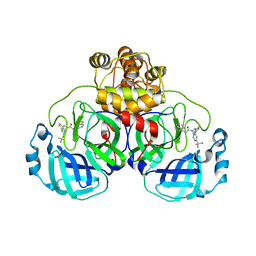 | | Crystal structure of SARS-CoV-2 Mpro with compound C3 | | Descriptor: | 3C-like proteinase, N-(4-tert-butylphenyl)-N-[(4-chlorothiophen-2-yl)methyl]-2-(isoquinolin-4-yl)acetamide | | Authors: | Worrall, L.J, Lee, J, Strynadka, N.C.J. | | Deposit date: | 2022-05-24 | | Release date: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A novel class of broad-spectrum active-site-directed 3C-like protease inhibitors with nanomolar antiviral activity against highly immune-evasive SARS-CoV-2 Omicron subvariants.
Emerg Microbes Infect, 12, 2023
|
|
1S2M
 
 | |
1QG1
 
 | |
1GUR
 
 | | GURMARIN, A SWEET TASTE-SUPPRESSING POLYPEPTIDE, NMR, 10 STRUCTURES | | Descriptor: | GURMARIN | | Authors: | Arai, K, Ishima, R, Morikawa, S, Imoto, T, Yoshimura, S, Aimoto, S, Akasaka, K. | | Deposit date: | 1996-03-12 | | Release date: | 1996-08-01 | | Last modified: | 2019-12-25 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of gurmarin, a sweet taste-suppressing polypeptide.
J.Biomol.NMR, 5, 1995
|
|
1HQO
 
 | | CRYSTAL STRUCTURE OF THE NITROGEN REGULATION FRAGMENT OF THE YEAST PRION PROTEIN URE2P | | Descriptor: | URE2 PROTEIN | | Authors: | Umland, T.C, Taylor, K.L, Rhee, S, Wickner, R.B, Davies, D.R. | | Deposit date: | 2000-12-18 | | Release date: | 2001-02-14 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystal structure of the nitrogen regulation fragment of the yeast prion protein Ure2p.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1GPS
 
 | | SOLUTION STRUCTURE OF GAMMA 1-H AND GAMMA 1-P THIONINS FROM BARLEY AND WHEAT ENDOSPERM DETERMINED BY 1H-NMR: A STRUCTURAL MOTIF COMMON TO TOXIC ARTHROPOD PROTEINS | | Descriptor: | GAMMA-1-P THIONIN | | Authors: | Bruix, M, Jimenez, M.A, Santoro, J, Gonzalez, C, Colilla, F.J, Mendez, E, Rico, M. | | Deposit date: | 1992-07-29 | | Release date: | 1993-10-31 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of gamma 1-H and gamma 1-P thionins from barley and wheat endosperm determined by 1H-NMR: a structural motif common to toxic arthropod proteins.
Biochemistry, 32, 1993
|
|
1FY3
 
 | | [G175Q]HBP, A mutant of human heparin binding protein (CAP37) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Kastrup, J.S, Linde, V, Pedersen, A.K, Stoffer, B, Iversen, L.F, Larsen, I.K, Rasmussen, P.B, Flodgaard, H.J, Bjorn, S.E. | | Deposit date: | 2000-09-28 | | Release date: | 2001-09-28 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Two mutants of human heparin binding protein (CAP37): toward the understanding of the nature of lipid A/LPS and BPTI binding.
Proteins, 42, 2001
|
|
1HNR
 
 | | H-NS (DNA-BINDING DOMAIN) | | Descriptor: | H-NS | | Authors: | Shindo, H, Iwaki, T, Ieda, R, Kurumizaka, H, Ueguchi, C, Mizuno, T, Morikawa, S, Nakamura, H, Kuboniwa, H. | | Deposit date: | 1995-04-06 | | Release date: | 1995-07-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the DNA binding domain of a nucleoid-associated protein, H-NS, from Escherichia coli.
FEBS Lett., 360, 1995
|
|
1IRH
 
 | | The Solution Structure of The Third Kunitz Domain of Tissue Factor Pathway Inhibitor | | Descriptor: | tissue factor pathway inhibitor | | Authors: | Mine, S, Yamazaki, T, Miyata, T, Hara, S, Kato, H. | | Deposit date: | 2001-10-02 | | Release date: | 2002-02-06 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structural mechanism for heparin-binding of the third Kunitz domain of human tissue factor pathway inhibitor.
Biochemistry, 41, 2002
|
|
1RHO
 
 | | STRUCTURE OF RHO GUANINE NUCLEOTIDE DISSOCIATION INHIBITOR | | Descriptor: | RHO GDP-DISSOCIATION INHIBITOR 1, SULFATE ION | | Authors: | Keep, N.H, Moody, P.C.E, Roberts, G.C.K. | | Deposit date: | 1996-10-12 | | Release date: | 1997-10-15 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A modulator of rho family G proteins, rhoGDI, binds these G proteins via an immunoglobulin-like domain and a flexible N-terminal arm.
Structure, 5, 1997
|
|
1FY1
 
 | | [R23S,F25E]HBP, A MUTANT OF HUMAN HEPARIN BINDING PROTEIN (CAP37) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ETHANOL, ... | | Authors: | Kastrup, J.S, Linde, V, Pedersen, A.K, Stoffer, B, Iversen, L.F, Larsen, I.K, Rasmussen, P.B, Flodgaard, H.J, Bjorn, S.E. | | Deposit date: | 2000-09-28 | | Release date: | 2001-09-28 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Two mutants of human heparin binding protein (CAP37): toward the understanding of the nature of lipid A/LPS and BPTI binding.
Proteins, 42, 2001
|
|
1TAP
 
 | | NMR SOLUTION STRUCTURE OF RECOMBINANT TICK ANTICOAGULANT PROTEIN (RTAP), A FACTOR XA INHIBITOR FROM THE TICK ORNITHODOROS MOUBATA | | Descriptor: | FACTOR XA INHIBITOR | | Authors: | Antuch, W, Guntert, P, Billeter, M, Wuthrich, K. | | Deposit date: | 1994-08-16 | | Release date: | 1994-11-30 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of the recombinant tick anticoagulant protein (rTAP), a factor Xa inhibitor from the tick Ornithodoros moubata.
FEBS Lett., 352, 1994
|
|
1PCA
 
 | | THREE DIMENSIONAL STRUCTURE OF PORCINE PANCREATIC PROCARBOXYPEPTIDASE A. A COMPARISON OF THE A AND B ZYMOGENS AND THEIR DETERMINANTS FOR INHIBITION AND ACTIVATION | | Descriptor: | CITRIC ACID, PROCARBOXYPEPTIDASE A PCPA, VALINE, ... | | Authors: | Guasch, A, Coll, M, Aviles, F.X, Huber, R. | | Deposit date: | 1991-10-28 | | Release date: | 1993-10-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Three-dimensional structure of porcine pancreatic procarboxypeptidase A. A comparison of the A and B zymogens and their determinants for inhibition and activation.
J.Mol.Biol., 224, 1992
|
|
1SM7
 
 | | Solution structure of the recombinant pronapin precursor, BnIb. | | Descriptor: | recombinant Ib pronapin | | Authors: | Pantoja-Uceda, D, Palomares, O, Bruix, M, Villalba, M, Rodriguez, R, Rico, M, Santoro, J. | | Deposit date: | 2004-03-08 | | Release date: | 2005-02-01 | | Last modified: | 2013-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure and stability against digestion of rproBnIb, a recombinant 2S albumin from rapeseed: relationship to its allergenic properties.
Biochemistry, 43, 2004
|
|
1KTH
 
 | |
1UEO
 
 | | Solution structure of the [T8A]-Penaeidin-3 | | Descriptor: | Penaeidin-3a | | Authors: | Yang, Y, Poncet, J, Garnier, J, Zatylny, C, Bachere, E, Aumelas, A. | | Deposit date: | 2003-05-20 | | Release date: | 2003-10-21 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the recombinant penaeidin-3, a shrimp antimicrobial peptide
J.Biol.Chem., 278, 2003
|
|
6NY0
 
 | |
8HEF
 
 | | The Crystal structure of deuterated S-217622 (Ensitrelvir) bound to the main protease (3CLpro/Mpro) of SARS-CoV-2 | | Descriptor: | 3C-like proteinase, 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione, GLYCEROL | | Authors: | Yan, M, Zhang, H. | | Deposit date: | 2022-11-08 | | Release date: | 2023-04-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Synthesis of deuterated S-217622 (Ensitrelvir) with antiviral activity against coronaviruses including SARS-CoV-2.
Antiviral Res., 213, 2023
|
|
6N8B
 
 | | Crystal structure of transcription regulator AcaB from uropathogenic E. coli | | Descriptor: | CALCIUM ION, transcription regulator AcaB | | Authors: | Luo, Z, Hancock, S.J, Schembri, M.A, Kobe, B. | | Deposit date: | 2018-11-29 | | Release date: | 2020-07-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Comprehensive analysis of IncC plasmid conjugation identifies a crucial role for the transcriptional regulator AcaB.
Nat Microbiol, 5, 2020
|
|
6N8A
 
 | | Crystal structure of selenomethionine-containing AcaB from uropathogenic E. coli | | Descriptor: | CHLORIDE ION, transcription regulator AcaB | | Authors: | Luo, Z, Hancock, S.J, Schembri, M.A, Kobe, B. | | Deposit date: | 2018-11-28 | | Release date: | 2020-07-15 | | Last modified: | 2021-01-27 | | Method: | X-RAY DIFFRACTION (3.4011 Å) | | Cite: | Comprehensive analysis of IncC plasmid conjugation identifies a crucial role for the transcriptional regulator AcaB.
Nat Microbiol, 5, 2020
|
|
7TOB
 
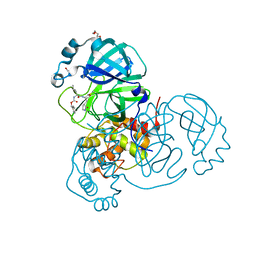 | | Crystal structure of the SARS-CoV-2 Omicron main protease (Mpro) in complex with inhibitor GC376 | | Descriptor: | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5, DI(HYDROXYETHYL)ETHER | | Authors: | Sacco, M.D, Wang, J, Chen, Y. | | Deposit date: | 2022-01-24 | | Release date: | 2022-02-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The P132H mutation in the main protease of Omicron SARS-CoV-2 decreases thermal stability without compromising catalysis or small-molecule drug inhibition.
Cell Res., 32, 2022
|
|
1AVA
 
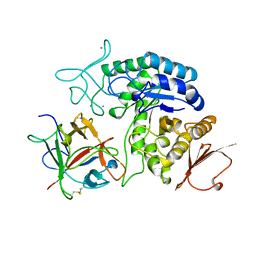 | | AMY2/BASI PROTEIN-PROTEIN COMPLEX FROM BARLEY SEED | | Descriptor: | BARLEY ALPHA-AMYLASE 2(CV MENUET), BARLEY ALPHA-AMYLASE/SUBTILISIN INHIBITOR, CALCIUM ION | | Authors: | Vallee, F, Kadziola, A, Bourne, Y, Juy, M, Svensson, B, Haser, R. | | Deposit date: | 1997-09-15 | | Release date: | 1999-03-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Barley alpha-amylase bound to its endogenous protein inhibitor BASI: crystal structure of the complex at 1.9 A resolution.
Structure, 6, 1998
|
|
1BAR
 
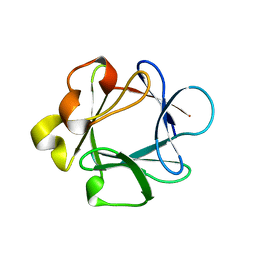 | | THREE-DIMENSIONAL STRUCTURES OF ACIDIC AND BASIC FIBROBLAST GROWTH FACTORS | | Descriptor: | ACIDIC FIBROBLAST GROWTH FACTOR | | Authors: | Zhu, X, Komiya, H, Chirino, A, Faham, S, Fox, G.M, Arakawa, T, Hsu, B.T, Rees, D.C. | | Deposit date: | 1992-09-29 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Three-dimensional structures of acidic and basic fibroblast growth factors.
Science, 251, 1991
|
|
1BAS
 
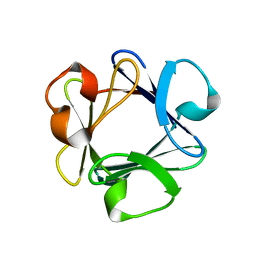 | |
