5I4U
 
 | | The crystal structure of PI3Kdelta with compound 34 | | Descriptor: | 2,4-diamino-6-{[(1S)-1-(5-chloro-4-oxo-3-phenyl-3,4-dihydroquinazolin-2-yl)ethyl]amino}pyrimidine-5-carbonitrile, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform | | Authors: | Somoza, J.R, Villasenor, A.G. | | Deposit date: | 2016-02-12 | | Release date: | 2017-02-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.372 Å) | | Cite: | The Design and Synthesis of Potent, Selective and Metabolically Stable PI3K[delta] Inhibitors
To be published
|
|
5VAL
 
 | | BRAF in Complex with N-(3-(tert-butyl)phenyl)-4-methyl-3-(6-morpholinopyrimidin-4-yl)benzamide | | Descriptor: | N-(3-tert-butylphenyl)-4-methyl-3-[6-(morpholin-4-yl)pyrimidin-4-yl]benzamide, Serine/threonine-protein kinase B-raf | | Authors: | Mamo, M, Appleton, B.A. | | Deposit date: | 2017-03-27 | | Release date: | 2017-06-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Design and Discovery of N-(2-Methyl-5'-morpholino-6'-((tetrahydro-2H-pyran-4-yl)oxy)-[3,3'-bipyridin]-5-yl)-3-(trifluoromethyl)benzamide (RAF709): A Potent, Selective, and Efficacious RAF Inhibitor Targeting RAS Mutant Cancers.
J. Med. Chem., 60, 2017
|
|
8VXS
 
 | | UIC-15-BPE extension of UIC-1 | | Descriptor: | ACETONITRILE, UIC-15-BPE extension of UIC-1 | | Authors: | Ganatra, P. | | Deposit date: | 2024-02-06 | | Release date: | 2024-08-07 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Diverse Proteomimetic Frameworks via Rational Design of pi-Stacking Peptide Tectons.
J.Am.Chem.Soc., 146, 2024
|
|
6VPA
 
 | |
5VAS
 
 | | Pekin duck egg lysozyme isoform III (DEL-III), orthorhombic form | | Descriptor: | GLYCEROL, Lysozyme, PHOSPHATE ION | | Authors: | Christie, M, Christ, D, Langley, D.B. | | Deposit date: | 2017-03-27 | | Release date: | 2018-04-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural basis of antigen recognition: crystal structure of duck egg lysozyme.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
8VCA
 
 | | Crystal Structure of plant Carboxylesterase 15 | | Descriptor: | Carboxylesterase 15, GLYCEROL | | Authors: | Palayam, M, Shabek, N. | | Deposit date: | 2023-12-13 | | Release date: | 2024-08-07 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insights into strigolactone catabolism by carboxylesterases reveal a conserved conformational regulation.
Nat Commun, 15, 2024
|
|
6VKE
 
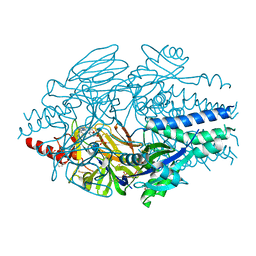 | | Crystal Structure of Inhibitor JNJ-40012665 in Complex with Prefusion RSV F Glycoprotein | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, 4-(5-chloro-2-{[1-(3,4-dimethoxyphenyl)-2-oxo-1,2-dihydro-3H-imidazo[4,5-c]pyridin-3-yl]methyl}-1H-indol-1-yl)butanenitrile, CHLORIDE ION, ... | | Authors: | McLellan, J.S. | | Deposit date: | 2020-01-20 | | Release date: | 2020-05-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery of 3-({5-Chloro-1-[3-(methylsulfonyl)propyl]-1H-indol-2-yl}methyl)-1-(2,2,2-trifluoroethyl)-1,3-dihydro-2H-imidazo[4,5-c]pyridin-2-one (JNJ-53718678), a Potent and Orally Bioavailable Fusion Inhibitor of Respiratory Syncytial Virus.
J.Med.Chem., 63, 2020
|
|
6VKQ
 
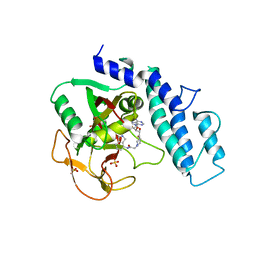 | | Crystal Structure of human PARP-1 CAT domain bound to inhibitor EB-47 | | Descriptor: | 2-[4-[(2S,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]carbonylpiperazin-1-yl]-N-(1-oxidanylidene-2,3-dihydroisoindol-4-yl)ethanamide, Poly [ADP-ribose] polymerase 1, SULFATE ION | | Authors: | Steffen, J.D, Pascal, J.M. | | Deposit date: | 2020-01-21 | | Release date: | 2020-06-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for allosteric PARP-1 retention on DNA breaks.
Science, 368, 2020
|
|
7LTT
 
 | | SAMHD1(113-626) H206R D207N R366C | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, MAGNESIUM ION | | Authors: | Temple, J.T, Bowen, N.E. | | Deposit date: | 2021-02-20 | | Release date: | 2021-11-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and functional characterization explains loss of dNTPase activity of the cancer-specific R366C/H mutant SAMHD1 proteins.
J.Biol.Chem., 297, 2021
|
|
5IIP
 
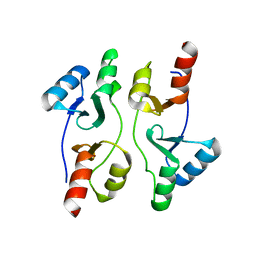 | | Staphylococcus aureus OpuCA | | Descriptor: | Glycine betaine/carnitine/choline ABC transporter%2C ATP-binding protein%2C putative | | Authors: | Tosi, T, Campeotto, I, Freemont, P.S, Grundling, A. | | Deposit date: | 2016-03-01 | | Release date: | 2016-08-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The second messenger c-di-AMP inhibits the osmolyte uptake system OpuC in Staphylococcus aureus.
Sci.Signal., 9, 2016
|
|
5I5R
 
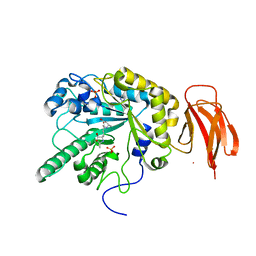 | | Crystal structure of a bacterial fucosidase with iminocyclitol (2S,3S,4R,5S)-3,4-dihydroxy-2-ethynyl-5-methylpyrrolidine | | Descriptor: | (2S,3S,4R,5S,2'S,3'S,4'R,5'S)-2,2'-(butane-1,4-diyl)bis(5-methylpyrrolidine-3,4-diol), Alpha-L-fucosidase, IMIDAZOLE, ... | | Authors: | Wright, D.W, Davies, G.J. | | Deposit date: | 2016-02-15 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Will be disclosed later
To Be Published
|
|
6VPG
 
 | |
8VFZ
 
 | |
6VPM
 
 | |
5V97
 
 | |
5I6L
 
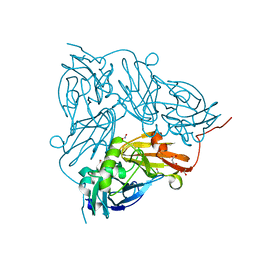 | | Crystal Structure of Copper Nitrite Reductase at 100K after 2.76 MGy | | Descriptor: | ACETATE ION, COPPER (II) ION, Copper-containing nitrite reductase, ... | | Authors: | Horrell, S, Hough, M.A, Strange, R.W. | | Deposit date: | 2016-02-16 | | Release date: | 2016-07-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Serial crystallography captures enzyme catalysis in copper nitrite reductase at atomic resolution from one crystal.
Iucrj, 3, 2016
|
|
8VZV
 
 | | Human TDO (hTDO) in complex with LM10 | | Descriptor: | 6-fluoro-3-[(E)-2-(1H-tetrazol-5-yl)ethenyl]-1H-indole, PROTOPORPHYRIN IX CONTAINING FE, Tryptophan 2,3-dioxygenase, ... | | Authors: | Ishigami, I, Yeh, S.-R, Lewis-Ballester, A. | | Deposit date: | 2024-02-12 | | Release date: | 2024-08-28 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structural Insights into Protein-Inhibitor Interactions in Human Tryptophan Dioxygenase.
J.Med.Chem., 67, 2024
|
|
8VBL
 
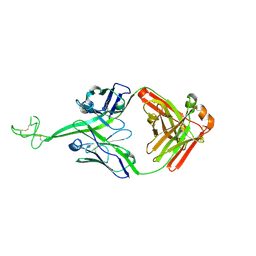 | | Structure of bovine anti-HIV Fab ElsE6 | | Descriptor: | Bovine Fab ElsE6 heavy chain, Bovine Fab ElsE6 light chain | | Authors: | Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2023-12-12 | | Release date: | 2024-09-04 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Immunization of cows with HIV envelope trimers generates broadly neutralizing antibodies to the V2-apex from the ultralong CDRH3 repertoire.
Plos Pathog., 20, 2024
|
|
5I6T
 
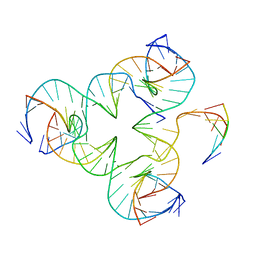 | | Crystal structure of color device state C | | Descriptor: | DNA (26-MER), DNA (5'-D(*AP*CP*AP*GP*TP*CP*GP*TP*GP*GP*TP*AP*TP*C)-3'), DNA (5'-D(*CP*AP*GP*AP*TP*AP*CP*CP*TP*GP*AP*TP*CP*GP*GP*AP*CP*TP*AP*CP*G)-3'), ... | | Authors: | Hao, Y, Birktoft, J, Seeman, N.C. | | Deposit date: | 2016-02-16 | | Release date: | 2017-01-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (5.283 Å) | | Cite: | A device that operates within a self-assembled 3D DNA crystal.
Nat Chem, 9, 2017
|
|
8VOF
 
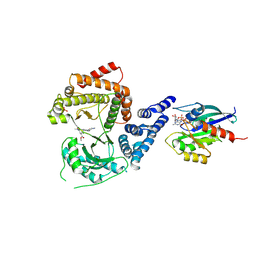 | | GI targeted CpPI4K inhibitor | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Isoform 2 of Phosphatidylinositol 4-kinase beta,Isoform 2 of Phosphatidylinositol 4-kinase beta,Phosphatidylinositol 4-kinase beta, Ras-related protein Rab-11A, ... | | Authors: | Knapp, M.S, Cuellar, C, Mamo, M. | | Deposit date: | 2024-01-15 | | Release date: | 2024-09-04 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Cryptosporidium PI(4)K inhibitor EDI048 is a gut-restricted parasiticidal agent to treat paediatric enteric cryptosporidiosis.
Nat Microbiol, 9, 2024
|
|
8VOM
 
 | |
8VT8
 
 | | UIC-13-BIF-A4Dab NBD-Cl binding | | Descriptor: | 4-chloro-7-nitrobenzofurazan, ACETONITRILE, UIC-13-BIF-A4Dab-NBD-Cl | | Authors: | Ganatra, P. | | Deposit date: | 2024-01-26 | | Release date: | 2024-08-07 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Diverse Proteomimetic Frameworks via Rational Design of pi-Stacking Peptide Tectons.
J.Am.Chem.Soc., 146, 2024
|
|
5VAD
 
 | | Crystal structure of human Prolyl-tRNA synthetase (PRS) in complex with inhibitor | | Descriptor: | 3-[(cyclohexanecarbonyl)amino]-N-(2,3-dihydro-1H-inden-2-yl)pyrazine-2-carboxamide, Bifunctional glutamate/proline--tRNA ligase, PROLINE, ... | | Authors: | Okada, K, Skene, R.J. | | Deposit date: | 2017-03-24 | | Release date: | 2017-05-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Discovery of a novel prolyl-tRNA synthetase inhibitor and elucidation of its binding mode to the ATP site in complex with l-proline.
Biochem. Biophys. Res. Commun., 488, 2017
|
|
6VJP
 
 | | Structure of Staphylococcus aureus peptidoglycan O-acetyltransferase A (OatA) C-terminal catalytic domain | | Descriptor: | Acetyltransferase, SODIUM ION | | Authors: | Jones, C.J, Sychantha, D, Howell, P.L, Clarke, A.J. | | Deposit date: | 2020-01-16 | | Release date: | 2020-05-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.711 Å) | | Cite: | Structural basis for theO-acetyltransferase function of the extracytoplasmic domain of OatA fromStaphylococcus aureus.
J.Biol.Chem., 295, 2020
|
|
8VMG
 
 | | Crystal structure of GSK-3 26-383 bound to Axin 383-435 | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Enos, M.D, Gavagan, M, Jameson, N, Zalatan, J.G, Weis, W.I. | | Deposit date: | 2024-01-13 | | Release date: | 2024-08-28 | | Last modified: | 2024-09-25 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural and functional effects of phosphopriming and scaffolding in the kinase GSK-3 beta.
Sci.Signal., 17, 2024
|
|
