8PSY
 
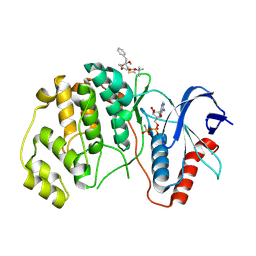 | | ERK2 covelently bound to RU68 cyclohexenone based inhibitor | | Descriptor: | AMP PHOSPHORAMIDATE, Mitogen-activated protein kinase 1, ~{O}3-~{tert}-butyl ~{O}1-methyl (1~{S},3~{R})-4-oxidanylidene-1-(phenylmethyl)cyclohexane-1,3-dicarboxylate | | Authors: | Sok, P, Poti, A, Remenyi, A, Gogl, G. | | Deposit date: | 2023-07-13 | | Release date: | 2024-07-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Targeting a key protein-protein interaction surface on mitogen-activated protein kinases by a precision-guided warhead scaffold.
Nat Commun, 15, 2024
|
|
8PSW
 
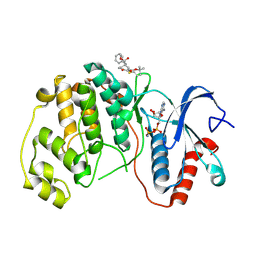 | | ERK2 covalently bound to RU67 cyclohexenone based inhibitor | | Descriptor: | AMP PHOSPHORAMIDATE, Mitogen-activated protein kinase 1, ~{O}3-~{tert}-butyl ~{O}1-methyl (1~{R},3~{R})-4-oxidanylidene-1-(phenylmethyl)cyclohexane-1,3-dicarboxylate | | Authors: | Sok, P, Poti, A, Remenyi, A, Gogl, G. | | Deposit date: | 2023-07-13 | | Release date: | 2024-07-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Targeting a key protein-protein interaction surface on mitogen-activated protein kinases by a precision-guided warhead scaffold.
Nat Commun, 15, 2024
|
|
8PST
 
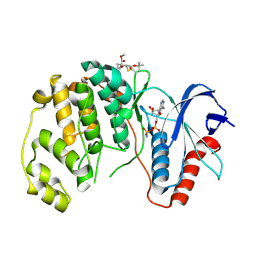 | | ERK2 covelently bound to RU60 cyclohexenone based inhibitor | | Descriptor: | AMP PHOSPHORAMIDATE, Mitogen-activated protein kinase 1, ~{O}3-~{tert}-butyl ~{O}1-methyl (1~{R},3~{S})-1-methyl-4-oxidanylidene-cyclohexane-1,3-dicarboxylate | | Authors: | Sok, P, Poti, A, Remenyi, A, Gogl, G. | | Deposit date: | 2023-07-13 | | Release date: | 2024-07-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Targeting a key protein-protein interaction surface on mitogen-activated protein kinases by a precision-guided warhead scaffold.
Nat Commun, 15, 2024
|
|
9CUA
 
 | |
9E4W
 
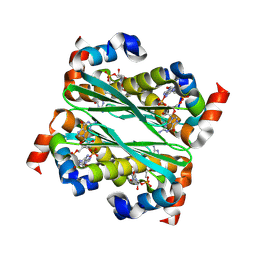 | | Structure of Bacillus phage SPO1 anti-CBASS 4 (Acb4) in complex with 3'3'-cGAMP | | Descriptor: | 2-amino-9-[(2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-9-(6-amino-9H-purin-9-yl)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecin-2-yl]-1,9-dihydro-6H-purin-6-one, anti-CBASS 4 (Acb4) | | Authors: | Chang, R.B, Toyoda, H.C, Hobbs, S.J, Richmond-Buccola, D, Wein, T, Burger, N, Chouchani, E.T, Sorek, R, Kranzusch, P.J. | | Deposit date: | 2024-10-25 | | Release date: | 2025-01-29 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | A widespread family of viral sponge proteins reveals specific inhibition of nucleotide signals in anti-phage defense.
Biorxiv, 2024
|
|
1MY2
 
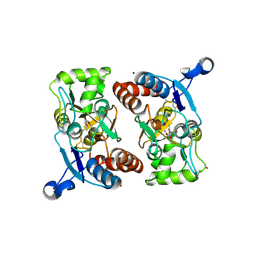 | | crystal titration experiment (AMPA complex control) | | Descriptor: | (S)-ALPHA-AMINO-3-HYDROXY-5-METHYL-4-ISOXAZOLEPROPIONIC ACID, GLUTAMATE RECEPTOR 2, ZINC ION | | Authors: | Jin, R, Gouaux, E. | | Deposit date: | 2002-10-03 | | Release date: | 2003-06-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Probing the function, conformational plasticity, and dimer-dimer contacts of the GluR2 ligand-binding core: studies of 5-substituted willardiines and GluR2 S1S2 in the crystal.
Biochemistry, 42, 2003
|
|
8QVZ
 
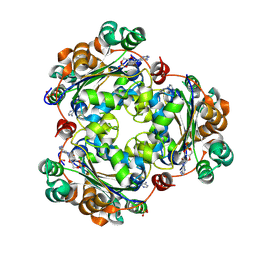 | | Human NDPK-C in complex with cAMP | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Nucleoside diphosphate kinase 3, SULFATE ION | | Authors: | Werten, S, Amjadi, R, Scheffzek, K. | | Deposit date: | 2023-10-18 | | Release date: | 2024-09-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.774 Å) | | Cite: | Mechanistic Insights into Substrate Recognition of Human Nucleoside Diphosphate Kinase C Based on Nucleotide-Induced Structural Changes.
Int J Mol Sci, 25, 2024
|
|
7MP9
 
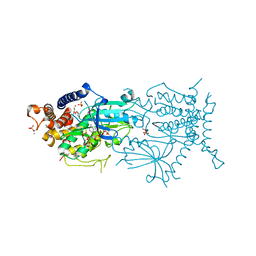 | | Crystal structure of the cytosolic domain of Tribolium castaneum PINK1 phosphorylated at Ser205 in complex with ADP analog | | Descriptor: | AMP PHOSPHORAMIDATE, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Rasool, S, Veyron, S, Trempe, J.F. | | Deposit date: | 2021-05-04 | | Release date: | 2021-12-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mechanism of PINK1 activation by autophosphorylation and insights into assembly on the TOM complex.
Mol.Cell, 82, 2022
|
|
2UXA
 
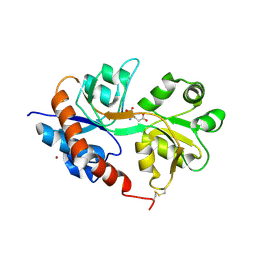 | | Crystal structure of the GluR2-flip ligand binding domain, r/g unedited. | | Descriptor: | GLUTAMATE RECEPTOR SUBUNIT GLUR2-FLIP, GLUTAMIC ACID, ZINC ION | | Authors: | Greger, I.H, Akamine, P, Khatri, L, Ziff, E.B. | | Deposit date: | 2007-03-27 | | Release date: | 2007-04-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Developmentally Regulated, Combinatorial RNA Processing Modulates Ampa Receptor Biogenesis.
Neuron, 51, 2006
|
|
4WB6
 
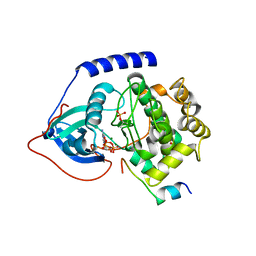 | | Crystal structure of a L205R mutant of human cAMP-dependent protein kinase A (catalytic alpha subunit) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, PKI (5-24), ... | | Authors: | Cheung, J, Ginter, C, Cassidy, M, Franklin, M.C, Rudolph, M.J, Hendrickson, W.A. | | Deposit date: | 2014-09-02 | | Release date: | 2015-01-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural insights into mis-regulation of protein kinase A in human tumors.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4WB5
 
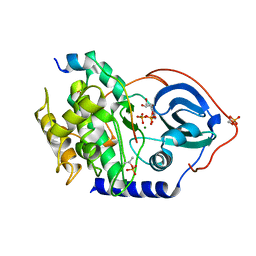 | | Crystal structure of human cAMP-dependent protein kinase A (catalytic alpha subunit) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, PKI (5-24), ... | | Authors: | Cheung, J, Ginter, C, Cassidy, M, Franklin, M.C, Rudolph, M.J, Hendrickson, W.A. | | Deposit date: | 2014-09-02 | | Release date: | 2015-01-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.641 Å) | | Cite: | Structural insights into mis-regulation of protein kinase A in human tumors.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
8T5Y
 
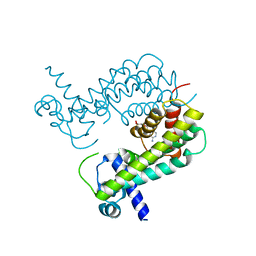 | |
7QV6
 
 | | Amyloid fibril from the antimicrobial peptide aurein 3.3 | | Descriptor: | Aurein-3.3 | | Authors: | Buecker, R, Seuring, C, Cazey, C, Veith, K, Garcia-Alai, M, Gruenewald, K, Landau, M. | | Deposit date: | 2022-01-19 | | Release date: | 2022-06-29 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The Cryo-EM structures of two amphibian antimicrobial cross-beta amyloid fibrils.
Nat Commun, 13, 2022
|
|
7QV5
 
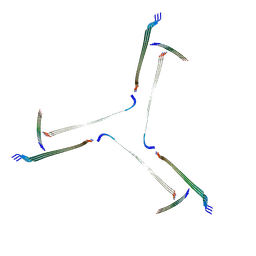 | | Amyloid fibril from the antimicrobial peptide uperin 3.5 | | Descriptor: | Uperin-3.5 | | Authors: | Buecker, R, Seuring, C, Cazey, C, Veith, K, Garcia-Alai, M, Gruenewald, K, Landau, M. | | Deposit date: | 2022-01-19 | | Release date: | 2022-06-29 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | The Cryo-EM structures of two amphibian antimicrobial cross-beta amyloid fibrils.
Nat Commun, 13, 2022
|
|
8WNG
 
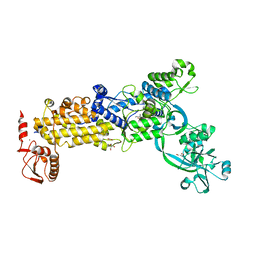 | | Crystal structure of H. pylori isoleucyl-tRNA synthetase (HpIleRS) in complex with Ile | | Descriptor: | ACETATE ION, GLYCEROL, ISOLEUCINE, ... | | Authors: | Guo, Y, Li, S, Zhang, T. | | Deposit date: | 2023-10-05 | | Release date: | 2024-02-14 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural basis for substrate and antibiotic recognition by Helicobacter pylori isoleucyl-tRNA synthetase.
Febs Lett., 598, 2024
|
|
3K8J
 
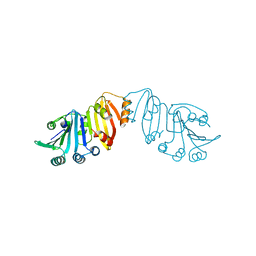 | | Structure of crystal form III of TP0453 | | Descriptor: | 30kLP | | Authors: | Zhu, G, Luthra, A, Desrosiers, D, Koszelak-Rosenblum, M, Mulay, V, Radolf, J.D, Malkowski, M.G. | | Deposit date: | 2009-10-14 | | Release date: | 2010-10-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Transition from Closed to Open Conformation of Treponema pallidum Outer Membrane-associated Lipoprotein TP0453 Involves Membrane Sensing and Integration by Two Amphipathic Helices.
J.Biol.Chem., 286, 2011
|
|
3K8H
 
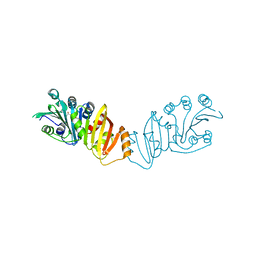 | | Structure of crystal form I of TP0453 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 30kLP | | Authors: | Zhu, G, Luthra, A, Desrosiers, D, Koszelak-Rosenblum, M, Mulay, V, Radolf, J.D, Malkowski, M.G. | | Deposit date: | 2009-10-14 | | Release date: | 2010-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | The Transition from Closed to Open Conformation of Treponema pallidum Outer Membrane-associated Lipoprotein TP0453 Involves Membrane Sensing and Integration by Two Amphipathic Helices.
J.Biol.Chem., 286, 2011
|
|
1REK
 
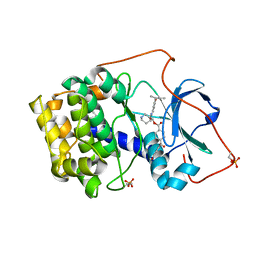 | | Crystal structure of cAMP-dependent protein kinase complexed with balanol analog 8 | | Descriptor: | 3-[(3-SEC-BUTYL-4-HYDROXYBENZOYL)AMINO]AZEPAN-4-YL 4-(2-HYDROXY-5-METHOXYBENZOYL)BENZOATE, PENTANAL, cAMP-dependent protein kinase, ... | | Authors: | Akamine, P, Madhusudan, Brunton, L.L, Ou, H.D, Canaves, J.M, Xuong, N.H, Taylor, S.S. | | Deposit date: | 2003-11-06 | | Release date: | 2004-02-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Balanol analogues probe specificity determinants and the conformational malleability of the cyclic 3',5'-adenosine monophosphate-dependent protein kinase catalytic subunit
Biochemistry, 43, 2004
|
|
1RE8
 
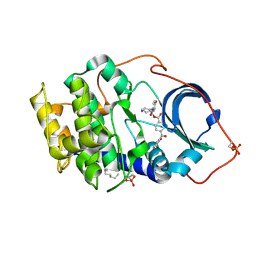 | | Crystal structure of cAMP-dependent protein kinase complexed with balanol analog 2 | | Descriptor: | 3-[(4-HYDROXYBENZOYL)AMINO]AZEPAN-4-YL 4-(2-HYDROXYBENZOYL)BENZOATE, N-OCTANE, cAMP-dependent protein kinase, ... | | Authors: | Akamine, P, Madhusudan, Brunton, L.L, Ou, H.D, Canaves, J.M, Xuong, N.H, Taylor, S.S. | | Deposit date: | 2003-11-06 | | Release date: | 2004-02-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Balanol analogues probe specificity determinants and the conformational malleability of the cyclic 3',5'-adenosine monophosphate-dependent protein kinase catalytic subunit
Biochemistry, 43, 2004
|
|
5SXS
 
 | |
1REJ
 
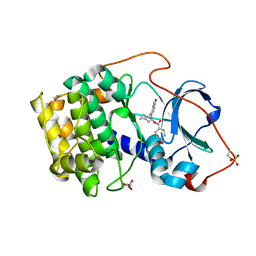 | | Crystal structure of cAMP-dependent protein kinase complexed with balanol analog 1 | | Descriptor: | 3-[(4-HYDROXYBENZOYL)AMINO]AZEPAN-4-YL 4-HYDROXYBENZOATE, cAMP-dependent protein kinase, alpha-catalytic subunit | | Authors: | Akamine, P, Madhusudan, Brunton, L.L, Ou, H.D, Canaves, J.M, Xuong, N.H, Taylor, S.S. | | Deposit date: | 2003-11-06 | | Release date: | 2004-02-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Balanol analogues probe specificity determinants and the conformational malleability of the cyclic 3',5'-adenosine monophosphate-dependent protein kinase catalytic subunit
Biochemistry, 43, 2004
|
|
5NC8
 
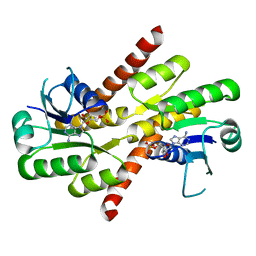 | | Shewanella denitrificans Kef CTD in AMP bound form | | Descriptor: | ADENOSINE MONOPHOSPHATE, Potassium efflux system protein | | Authors: | Pliotas, C, Naismith, J.H. | | Deposit date: | 2017-03-03 | | Release date: | 2017-04-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Adenosine Monophosphate Binding Stabilizes the KTN Domain of the Shewanella denitrificans Kef Potassium Efflux System.
Biochemistry, 56, 2017
|
|
8U97
 
 | |
4UQP
 
 | | High-resolution structure of the D. fructosovorans NiFe-hydrogenase L122A mutant after exposure to air | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE3-S4 CLUSTER, GLYCEROL, ... | | Authors: | Volbeda, A, Martin, L, Barbier, E, Gutierrez-Sanz, O, DeLacey, A.L, Liebgott, P.P, Dementin, S, Rousset, M, Fontecilla-Camps, J.C. | | Deposit date: | 2014-06-24 | | Release date: | 2014-10-29 | | Last modified: | 2023-03-15 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Crystallographic Studies of [Nife]-Hydrogenase Mutants: Towards Consensus Structures for the Elusive Unready Oxidized States.
J.Biol.Inorg.Chem., 20, 2015
|
|
4UQL
 
 | | High-resolution structure of a Ni-A Ni-Sox mixture of the D. fructosovorans NiFe-hydrogenase L122A mutant | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, CHLORIDE ION, FE3-S4 CLUSTER, ... | | Authors: | Volbeda, A, Martin, L, Barbier, E, Gutierrez-Sanz, O, DeLacey, A.L, Liebgott, P.P, Dementin, S, Rousset, M, Fontecilla-Camps, J.C. | | Deposit date: | 2014-06-24 | | Release date: | 2014-10-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Crystallographic Studies of [Nife]-Hydrogenase Mutants: Towards Consensus Structures for the Elusive Unready Oxidized States.
J.Biol.Inorg.Chem., 20, 2015
|
|
