8SVA
 
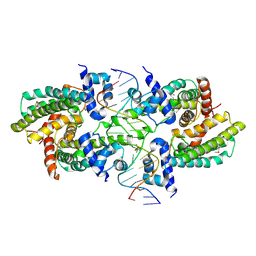 | | Structure of the Rhodococcus sp. USK13 DarR-20 bp DNA complex | | Descriptor: | DNA (5'-D(*TP*AP*GP*AP*TP*AP*CP*TP*CP*CP*GP*GP*AP*GP*TP*AP*TP*CP*TP*A)-3'), PHOSPHATE ION, TetR/AcrR family transcriptional regulator | | Authors: | Schumacher, M.A. | | Deposit date: | 2023-05-15 | | Release date: | 2023-11-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Structures of the DarR transcription regulator reveal unique modes of second messenger and DNA binding.
Nat Commun, 14, 2023
|
|
8SUA
 
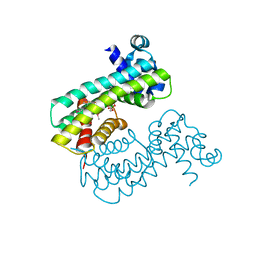 | | Structure of M. baixiangningiae DarR-ligand complex | | Descriptor: | 3-azanyl-3-(hydroxymethyl)-1,5,7,11-tetraoxa-6$l^{4}-boraspiro[5.5]undecan-9-ol, DarR | | Authors: | Schumacher, M.A. | | Deposit date: | 2023-05-11 | | Release date: | 2023-11-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structures of the DarR transcription regulator reveal unique modes of second messenger and DNA binding.
Nat Commun, 14, 2023
|
|
8SVD
 
 | |
8SLZ
 
 | |
8T3P
 
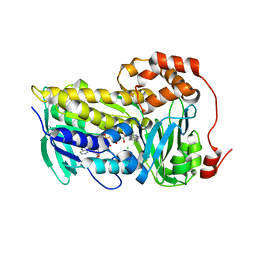 | |
8T1B
 
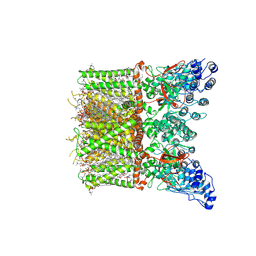 | | Cryo-EM structure of full-length human TRPV4 in apo state | | Descriptor: | (2R)-2-{[(4-O-hexopyranosyl-beta-D-glucopyranosyl)oxy]methyl}-4-{[(25R)-5beta,14beta,17beta-spirostan-3beta-yl]oxy}butyl 4-O-alpha-D-glucopyranosyl-beta-D-glucopyranoside, SODIUM ION, Transient receptor potential cation channel subfamily V member 4/Enhanced green fluorescent protein chimera, ... | | Authors: | Nadezhdin, K.D, Talyzina, I.A, Neuberger, A, Sobolevsky, A.I. | | Deposit date: | 2023-06-02 | | Release date: | 2023-07-05 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure of human TRPV4 in complex with GTPase RhoA.
Nat Commun, 14, 2023
|
|
8T1D
 
 | | Open-state cryo-EM structure of full-length human TRPV4 in complex with agonist 4a-PDD | | Descriptor: | (1aR,1bS,4aS,7aS,7bS,8R,9R,9aS)-9a-(decanoyloxy)-4a,7b-dihydroxy-3-(hydroxymethyl)-1,1,6,8-tetramethyl-5-oxo-1a,1b,4,4a,5,7a,7b,8,9,9a-decahydro-1H-cyclopropa[3,4]benzo[1,2-e]azulen-9-yl decanoate, Transient receptor potential cation channel subfamily V member 4/Enhanced green fluorescent protein chimera | | Authors: | Talyzina, I.A, Nadezhdin, K.D, Neuberger, A, Sobolevsky, A.I. | | Deposit date: | 2023-06-02 | | Release date: | 2023-07-05 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Structure of human TRPV4 in complex with GTPase RhoA.
Nat Commun, 14, 2023
|
|
8T1F
 
 | | Cryo-EM structure of full-length human TRPV4 in complex with antagonist HC-067047 | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-methyl-1-[3-(morpholin-4-yl)propyl]-5-phenyl-N-[3-(trifluoromethyl)phenyl]-1H-pyrrole-3-carboxamide, Transient receptor potential cation channel subfamily V member 4/Enhanced green fluorescent protein chimera | | Authors: | Talyzina, I.A, Nadezhdin, K.D, Neuberger, A, Sobolevsky, A.I. | | Deposit date: | 2023-06-02 | | Release date: | 2023-07-05 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Structure of human TRPV4 in complex with GTPase RhoA.
Nat Commun, 14, 2023
|
|
8T1C
 
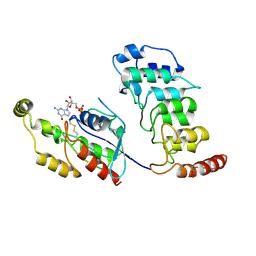 | | Cryo-EM structure of human TRPV4 ankyrin repeat domain in complex with GTPase RhoA | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Transforming protein RhoA, Transient receptor potential cation channel subfamily V member 4-Enhanced green fluorescent protein chimera | | Authors: | Nadezhdin, K.D, Talyzina, I.A, Neuberger, A, Sobolevsky, A.I. | | Deposit date: | 2023-06-02 | | Release date: | 2023-07-05 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Structure of human TRPV4 in complex with GTPase RhoA.
Nat Commun, 14, 2023
|
|
8T1E
 
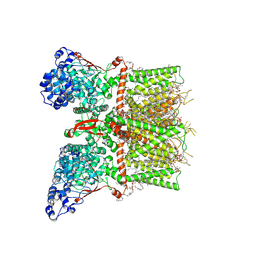 | | Closed-state cryo-EM structure of full-length human TRPV4 in the presence of 4a-PDD | | Descriptor: | (2R)-2-{[(4-O-hexopyranosyl-beta-D-glucopyranosyl)oxy]methyl}-4-{[(25R)-5beta,14beta,17beta-spirostan-3beta-yl]oxy}butyl 4-O-alpha-D-glucopyranosyl-beta-D-glucopyranoside, SODIUM ION, Transient receptor potential cation channel subfamily V member 4,Enhanced green fluorescent protein, ... | | Authors: | Talyzina, I.A, Nadezhdin, K.D, Neuberger, A, Sobolevsky, A.I. | | Deposit date: | 2023-06-02 | | Release date: | 2023-07-05 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Structure of human TRPV4 in complex with GTPase RhoA.
Nat Commun, 14, 2023
|
|
8T2U
 
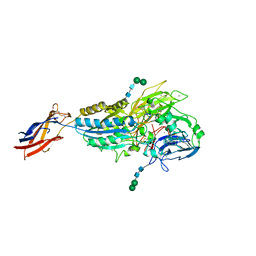 | | Cryo-EM Structures of Full-length Integrin alphaIIbbeta3 in Native Lipids complexed with Eptifibatide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Adair, B, Xiong, J.P, Yeager, M, Arnaout, M.A. | | Deposit date: | 2023-06-06 | | Release date: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structures of full-length integrin alpha IIb beta 3 in native lipids.
Nat Commun, 14, 2023
|
|
7SQ4
 
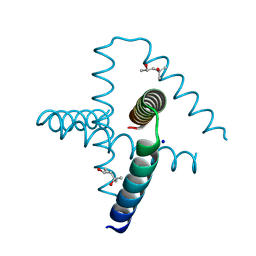 | | Designed trefoil knot protein, variant 2 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Designed trefoil knot protein, variant 2, ... | | Authors: | Takushi, B, Doyle, L, Stoddard, B.L, Bradley, P. | | Deposit date: | 2021-11-04 | | Release date: | 2022-06-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.493 Å) | | Cite: | De novo design of knotted tandem repeat proteins.
Nat Commun, 14, 2023
|
|
7SQ3
 
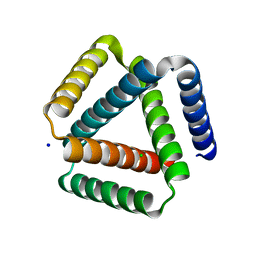 | | Designed trefoil knot protein, variant 1 | | Descriptor: | CHLORIDE ION, Designed trefoil knot protein, variant 1, ... | | Authors: | Takushi, B, Doyle, L, Stoddard, B.L, Bradley, P. | | Deposit date: | 2021-11-04 | | Release date: | 2022-06-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.453 Å) | | Cite: | De novo design of knotted tandem repeat proteins.
Nat Commun, 14, 2023
|
|
2NRY
 
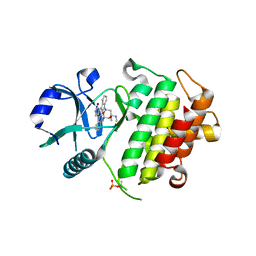 | | Crystal structure of IRAK-4 | | Descriptor: | STAUROSPORINE, interleukin-1 receptor-associated kinase 4 | | Authors: | Wang, Z, Liu, J, Walker, N.P.C. | | Deposit date: | 2006-11-02 | | Release date: | 2006-12-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structures of IRAK-4 kinase in complex with inhibitors: a serine/threonine kinase with tyrosine as a gatekeeper.
Structure, 14, 2006
|
|
7SJS
 
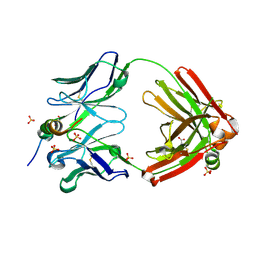 | |
8T1P
 
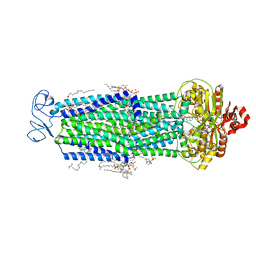 | |
8SZC
 
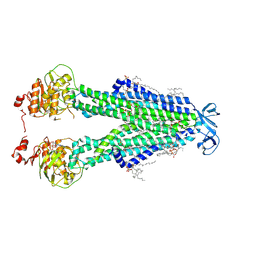 | |
8T3K
 
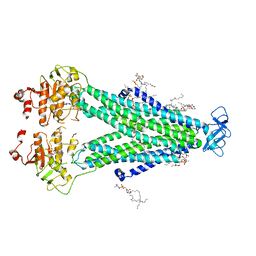 | |
7SU1
 
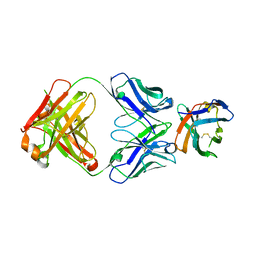 | | Crystal structure of an acidic pH-selective Ipilimumab variant Ipi.106 in complex with CTLA-4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Cytotoxic T-lymphocyte protein 4, Fab heavy chain, ... | | Authors: | Lee, P.S, Chau, B, Strop, P. | | Deposit date: | 2021-11-15 | | Release date: | 2022-03-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Improved therapeutic index of an acidic pH-selective antibody.
Mabs, 14, 2022
|
|
8QME
 
 | |
2OBD
 
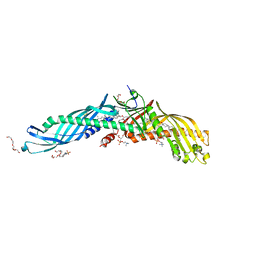 | | Crystal Structure of Cholesteryl Ester Transfer Protein | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Qiu, X. | | Deposit date: | 2006-12-18 | | Release date: | 2007-01-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of cholesteryl ester transfer protein reveals a long tunnel and four bound lipid molecules.
Nat.Struct.Mol.Biol., 14, 2007
|
|
8T7E
 
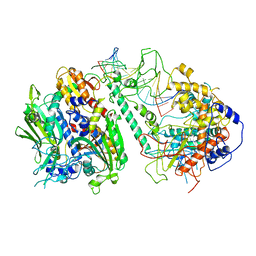 | | Cryo-EM structure of the Backtracking Initiation Complex (VII) of Human Mitochondrial DNA Polymerase Gamma | | Descriptor: | DNA polymerase subunit gamma-1, DNA polymerase subunit gamma-2, mitochondrial, ... | | Authors: | Nayak, A.R, Buchel, G, Herbine, K.H, Sarfallah, A, Sokolova, V.O, Zamudio-Ochoa, A, Temiakov, D. | | Deposit date: | 2023-06-20 | | Release date: | 2024-01-10 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Structural basis for DNA proofreading.
Nat Commun, 14, 2023
|
|
2OKK
 
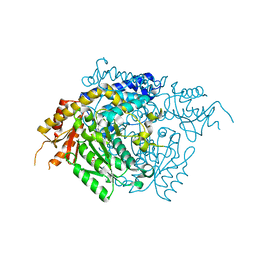 | | The X-ray crystal structure of the 65kDa isoform of Glutamic Acid Decarboxylase (GAD65) | | Descriptor: | GAMMA-AMINO-BUTANOIC ACID, GLYCEROL, Glutamate decarboxylase 2 | | Authors: | Buckle, A.M, Fenalti, G, Law, R.H.P, Whisstock, J.C. | | Deposit date: | 2007-01-17 | | Release date: | 2007-03-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | GABA production by glutamic acid decarboxylase is regulated by a dynamic catalytic loop.
Nat.Struct.Mol.Biol., 14, 2007
|
|
2NRU
 
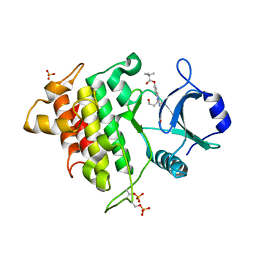 | | Crystal structure of IRAK-4 | | Descriptor: | 1-(3-HYDROXYPROPYL)-2-[(3-NITROBENZOYL)AMINO]-1H-BENZIMIDAZOL-5-YL PIVALATE, Interleukin-1 receptor-associated kinase 4, SULFATE ION | | Authors: | Wang, Z, Liu, J, Walker, N.P.C. | | Deposit date: | 2006-11-02 | | Release date: | 2006-12-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of IRAK-4 kinase in complex with inhibitors: a serine/threonine kinase with tyrosine as a gatekeeper.
Structure, 14, 2006
|
|
8RP4
 
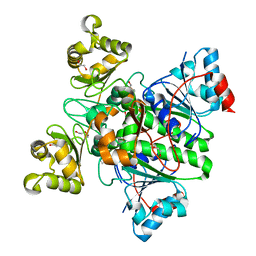 | | Alpha-Methylacyl-CoA racemase from Mycobacterium tuberculosis (D156A mutant) | | Descriptor: | 1,2-ETHANEDIOL, Alpha-methylacyl-CoA racemase, DI(HYDROXYETHYL)ETHER | | Authors: | Mojanaga, O.O, Acharya, K.R, Lloyd, M.D. | | Deposit date: | 2024-01-12 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | alpha-Methylacyl-CoA Racemase from Mycobacterium tuberculosis -Detailed Kinetic and Structural Characterization of the Active Site.
Biomolecules, 14, 2024
|
|
