2H32
 
 | | Crystal structure of the pre-B cell receptor | | Descriptor: | Immunoglobulin heavy chain, Immunoglobulin iota chain, Immunoglobulin omega chain, ... | | Authors: | Bankovich, A.J. | | Deposit date: | 2006-05-22 | | Release date: | 2007-04-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Insight into Pre-B Cell Receptor Function
Science, 316, 2007
|
|
3H3F
 
 | | Rabbit muscle L-lactate dehydrogenase in complex with NADH and oxamate | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ACETATE ION, L-lactate dehydrogenase A chain, ... | | Authors: | Bujacz, A, Bujacz, G, Swiderek, K, Paneth, P. | | Deposit date: | 2009-04-16 | | Release date: | 2009-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Modeling of isotope effects on binding oxamate to lactic dehydrogenase
J.Phys.Chem.B, 113, 2009
|
|
7JG2
 
 | | Secretory Immunoglobin A (SIgA) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Igh protein, ... | | Authors: | Kumar Bharathkar, S, Parker, B.P, Malyutin, A.G, Stadtmueller, B.M. | | Deposit date: | 2020-07-18 | | Release date: | 2020-11-11 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | The structures of Secretory and dimeric Immunoglobulin A.
Elife, 9, 2020
|
|
3H4P
 
 | | Proteasome 20S core particle from Methanocaldococcus jannaschii | | Descriptor: | Proteasome subunit alpha, Proteasome subunit beta | | Authors: | Jeffrey, P.D, Zhang, F, Hu, M, Tian, G, Zhang, P, Finley, D, Shi, Y. | | Deposit date: | 2009-04-20 | | Release date: | 2009-06-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | Structural Insights into the Regulatory Particle of the Proteasome from Methanocaldococcus jannaschii.
Mol.Cell, 34, 2009
|
|
2H5J
 
 | | Crystal strusture of caspase-3 with inhibitor Ac-DMQD-Cho | | Descriptor: | Ac-DMQD-Cho, caspase-3, p12 subunit, ... | | Authors: | Fang, B, Boross, P.I, Tozser, J, Weber, I.T. | | Deposit date: | 2006-05-26 | | Release date: | 2006-09-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and kinetic analysis of caspase-3 reveals role for s5 binding site in substrate recognition
J.Mol.Biol., 360, 2006
|
|
2H5Y
 
 | | Crystallographic structure of the Molybdate-Binding Protein of Xanthomonas citri at 1.7 Ang resolution bound to molybdate | | Descriptor: | MOLYBDATE ION, Molybdate-binding periplasmic protein, SULFATE ION | | Authors: | Balan, A, Santacruz, C.P, Ferreira, L.C.S, Barbosa, J.A.R.G. | | Deposit date: | 2006-05-29 | | Release date: | 2007-06-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystallization, data collection and phasing of the molybdate-binding protein of the phytopathogen Xanthomonas axonopodis pv. citri
Acta Crystallogr.,Sect.F, 62, 2006
|
|
2P87
 
 | | Crystal structure of the C-terminal domain of C. elegans pre-mRNA splicing factor Prp8 | | Descriptor: | Pre-mRNA-splicing factor Prp8 | | Authors: | Zhang, L, Shen, J, Guarnieri, M.T, Heroux, A, Yang, K, Zhao, R. | | Deposit date: | 2007-03-21 | | Release date: | 2007-05-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the C-terminal domain of splicing factor Prp8 carrying retinitis pigmentosa mutants
Protein Sci., 16, 2007
|
|
2H6G
 
 | |
7MDO
 
 | | Structure of human p97 ATPase L464P mutant | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Transitional endoplasmic reticulum ATPase | | Authors: | Zhang, X, Gui, L, Li, S, Nandi, P, Columbres, R.C, Wong, D.E, Moen, D.R, Lin, H.J, Chiu, P.-L, Chou, T.-F. | | Deposit date: | 2021-04-05 | | Release date: | 2021-08-25 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (4.12 Å) | | Cite: | Conserved L464 in p97 D1-D2 linker is critical for p97 cofactor regulated ATPase activity.
Biochem.J., 478, 2021
|
|
7MDM
 
 | | Structure of human p97 ATPase L464P mutant | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Transitional endoplasmic reticulum ATPase | | Authors: | Zhang, X, Gui, L, Li, S, Nandi, P, Columbres, R.C, Wong, D.E, Moen, D.R, Lin, H.J, Chiu, P.-L, Chou, T.-F. | | Deposit date: | 2021-04-05 | | Release date: | 2021-08-25 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (4.86 Å) | | Cite: | Conserved L464 in p97 D1-D2 linker is critical for p97 cofactor regulated ATPase activity.
Biochem.J., 478, 2021
|
|
2H7Q
 
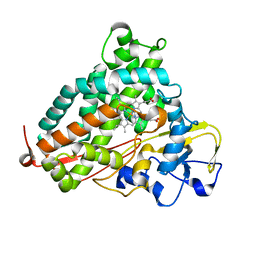 | | Cytochrome P450cam complexed with imidazole | | Descriptor: | Cytochrome P450-cam, IMIDAZOLE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Verras, A, Alian, A, Montellano, P.R. | | Deposit date: | 2006-06-02 | | Release date: | 2006-10-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Cytochrome P450 active site plasticity: attenuation of imidazole binding in cytochrome P450cam by an L244A mutation.
Protein Eng.Des.Sel., 19, 2006
|
|
3GLG
 
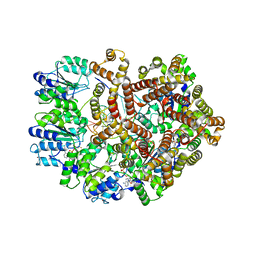 | | Crystal Structure of a Mutant (gammaT157A) E. coli Clamp Loader Bound to Primer-Template DNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, DNA (5'-D(*CP*TP*GP*GP*CP*CP*TP*AP*TP*A)-3'), ... | | Authors: | Simonetta, K.R, Seyedin, S.N, Kuriyan, J. | | Deposit date: | 2009-03-12 | | Release date: | 2009-05-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | The mechanism of ATP-dependent primer-template recognition by a clamp loader complex.
Cell(Cambridge,Mass.), 137, 2009
|
|
2H85
 
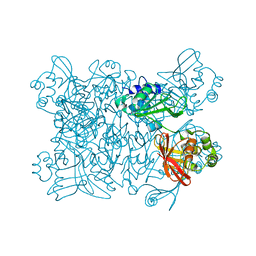 | | Crystal Structure of Nsp 15 from SARS | | Descriptor: | Putative orf1ab polyprotein | | Authors: | Ricagno, S, Egloff, M.P, Ulferts, R, Coutard, B, Nurizzo, D, Campanacci, V, Cambillau, C, Ziebuhr, J, Canard, B. | | Deposit date: | 2006-06-06 | | Release date: | 2006-08-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure and mechanistic determinants of SARS coronavirus nonstructural protein 15 define an endoribonuclease family.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
7JGO
 
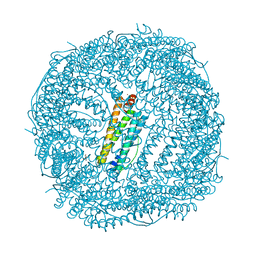 | | Crystal Structure of the Ni-bound Human Heavy-chain variant 122H-delta C-star with 2,5-furandihyrdoxamate collected at 278K | | Descriptor: | Ferritin heavy chain, NICKEL (II) ION, N~2~,N~5~-dihydroxyfuran-2,5-dicarboxamide, ... | | Authors: | Bailey, J.B, Tezcan, F.A. | | Deposit date: | 2020-07-19 | | Release date: | 2020-10-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.08 Å) | | Cite: | Tunable and Cooperative Thermomechanical Properties of Protein-Metal-Organic Frameworks.
J.Am.Chem.Soc., 142, 2020
|
|
7M20
 
 | | 18-mer HeLa-tubulin rings in complex with Cryptophycin 1 | | Descriptor: | Cryptophycin 1, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Eren, E. | | Deposit date: | 2021-03-15 | | Release date: | 2021-09-08 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.84 Å) | | Cite: | Conformational changes in tubulin upon binding cryptophycin-52 reveal its mechanism of action.
J.Biol.Chem., 297, 2021
|
|
3AWT
 
 | |
7MMN
 
 | |
3AWY
 
 | |
7JL2
 
 | | Cryo-EM structure of MDA5-dsRNA filament in complex with TRIM65 PSpry domain (Trimer) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Interferon-induced helicase C domain-containing protein 1, MAGNESIUM ION, ... | | Authors: | Kato, K, Ahmad, S, Hur, S. | | Deposit date: | 2020-07-29 | | Release date: | 2020-12-09 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural analysis of RIG-I-like receptors reveals ancient rules of engagement between diverse RNA helicases and TRIM ubiquitin ligases.
Mol.Cell, 81, 2021
|
|
3AWW
 
 | |
3H0S
 
 | |
3AWV
 
 | |
7MQ6
 
 | | Tetragonal Maltose Binding Protein in the presence of gold | | Descriptor: | CHLORIDE ION, GOLD ION, Maltodextrin-binding protein, ... | | Authors: | Thaker, A, Sirajudeen, L, Simmons, C.R, Nannenga, B.L. | | Deposit date: | 2021-05-05 | | Release date: | 2021-09-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.372 Å) | | Cite: | Structure-guided identification of a peptide for bio-enabled gold nanoparticle synthesis.
Biotechnol.Bioeng., 118, 2021
|
|
3AWS
 
 | |
7KOB
 
 | | Crystal structure of antigen 43b from Escherichia coli CFT073 | | Descriptor: | Antigen 43b, GLYCEROL | | Authors: | Martinez-Ortiz, G.C, Hor, L, Paxman, J.J, Heras, B. | | Deposit date: | 2020-11-07 | | Release date: | 2022-03-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Variation of Antigen 43 self-association modulates bacterial compacting within aggregates and biofilms.
NPJ Biofilms Microbiomes, 8, 2022
|
|
