7GTD
 
 | |
5NA7
 
 | | Structure of DPP III from Bacteroides thetaiotaomicron | | Descriptor: | CHLORIDE ION, Putative dipeptidyl-peptidase III, SULFATE ION, ... | | Authors: | Sabljic, I, Luic, M. | | Deposit date: | 2017-02-27 | | Release date: | 2017-11-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Crystal structure of dipeptidyl peptidase III from the human gut symbiont Bacteroides thetaiotaomicron.
PLoS ONE, 12, 2017
|
|
7GSU
 
 | |
7GTR
 
 | |
7GSK
 
 | |
7GTS
 
 | |
2Z9K
 
 | | Complex structure of SARS-CoV 3C-like protease with JMF1600 | | Descriptor: | (dimethylamino)(hydroxy)zinc', 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Lee, C.C, Wang, A.H. | | Deposit date: | 2007-09-20 | | Release date: | 2007-12-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis of mercury- and zinc-conjugated complexes as SARS-CoV 3C-like protease inhibitors.
Febs Lett., 581, 2007
|
|
7GTO
 
 | | PanDDA Analysis group deposition -- Crystal structure of PTP1B in complex with FMOOA000602a | | Descriptor: | (6aR,8R,12R,12aS)-5-methyl-5,6a,7,10,11,12a-hexahydro-6H,9H-pyrazolo[1',2':1,2]pyrazolo[4,3-c]quinolin-9-one, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Mehlman, T, Ginn, H.M, Keedy, D.A. | | Deposit date: | 2024-01-03 | | Release date: | 2024-01-24 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | An expanded view of ligandability in the allosteric enzyme PTP1B from computational reanalysis of large-scale crystallographic data.
Biorxiv, 2024
|
|
7GSR
 
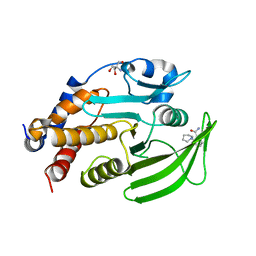 | |
5N1H
 
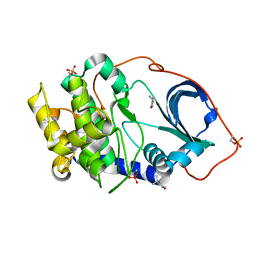 | |
7MIO
 
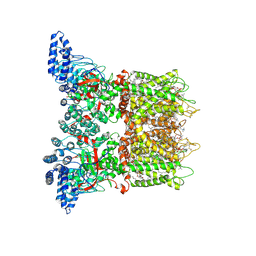 | | Mouse TRPV3 in cNW11 nanodiscs, open state at 42 degrees Celsius | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, SODIUM ION, Transient receptor potential cation channel subfamily V member 3 | | Authors: | Nadezhdin, K.D, Neuberger, A, Sobolevsky, A.I. | | Deposit date: | 2021-04-17 | | Release date: | 2021-07-21 | | Last modified: | 2021-07-28 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | Structural mechanism of heat-induced opening of a temperature-sensitive TRP channel.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7GS7
 
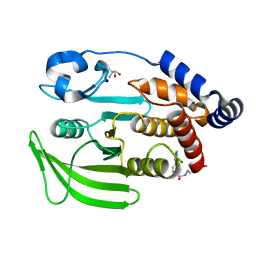 | | PanDDA Analysis group deposition -- Crystal structure of PTP1B in complex with FMOPL000621a | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-(1,2,3-thiadiazol-4-yl)phenyl ethylcarbamate, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Mehlman, T, Ginn, H.M, Keedy, D.A. | | Deposit date: | 2024-01-03 | | Release date: | 2024-01-24 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | An expanded view of ligandability in the allosteric enzyme PTP1B from computational reanalysis of large-scale crystallographic data.
Biorxiv, 2024
|
|
7KL9
 
 | |
7GSY
 
 | |
7GT5
 
 | | PanDDA Analysis group deposition -- Crystal structure of PTP1B in complex with FMOOA000529a | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Tyrosine-protein phosphatase non-receptor type 1, methyl [(3R,4S)-3-ethyl-4-hydroxy-1,1-dioxo-3,4-dihydro-1lambda~6~,2-benzothiazin-2(1H)-yl]acetate | | Authors: | Mehlman, T, Ginn, H.M, Keedy, D.A. | | Deposit date: | 2024-01-03 | | Release date: | 2024-01-24 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | An expanded view of ligandability in the allosteric enzyme PTP1B from computational reanalysis of large-scale crystallographic data.
Biorxiv, 2024
|
|
3KLF
 
 | | Crystal structure of wild-type HIV-1 Reverse Transcriptase crosslinked to a DSDNA with a bound excision product, AZTPPPPA | | Descriptor: | DNA (5'-D(*A*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)P*CP*GP*CP*CP*(2DA))-3'), DNA (5'-D(*AP*T*GP*CP*AP*TP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*GP*TP*G)-3'), GLYCEROL, ... | | Authors: | Tu, X, Das, K, Sarafianos, S.G, Arnold, E. | | Deposit date: | 2009-11-07 | | Release date: | 2010-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structural basis of HIV-1 resistance to AZT by excision.
Nat.Struct.Mol.Biol., 17, 2010
|
|
7ICF
 
 | |
5N27
 
 | |
7KRZ
 
 | | Human mitochondrial LONP1 in complex with Bortezomib | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Endogenous co-purified substrate, ... | | Authors: | Shin, M, Watson, E.R, Song, A.S, Mindrebo, J.T, Novick, S.R, Griffin, P, Wiseman, R.L, Lander, G.C. | | Deposit date: | 2020-11-20 | | Release date: | 2021-02-24 | | Last modified: | 2022-06-15 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structures of the human LONP1 protease reveal regulatory steps involved in protease activation.
Nat Commun, 12, 2021
|
|
7GTT
 
 | | PanDDA Analysis group deposition -- Crystal structure of PTP1B in complex with FMOPL000148a | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, N-(3,4-dihydroquinoline-1(2H)-carbothioyl)propanamide, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Mehlman, T, Ginn, H.M, Keedy, D.A. | | Deposit date: | 2024-01-03 | | Release date: | 2024-01-24 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | An expanded view of ligandability in the allosteric enzyme PTP1B from computational reanalysis of large-scale crystallographic data.
Biorxiv, 2024
|
|
7GSA
 
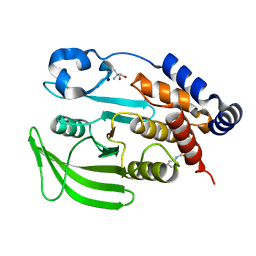 | |
7JGK
 
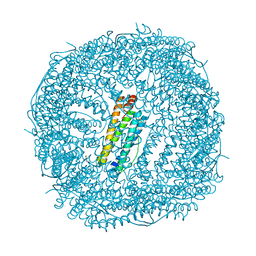 | | Crystal Structure of the Ni-bound Human Heavy-chain variant 122H-delta C-star with 2,5-furandihyrdoxamate collected at 100K | | Descriptor: | Ferritin heavy chain, NICKEL (II) ION, N~2~,N~5~-dihydroxyfuran-2,5-dicarboxamide, ... | | Authors: | Bailey, J.B, Tezcan, F.A. | | Deposit date: | 2020-07-19 | | Release date: | 2020-10-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Tunable and Cooperative Thermomechanical Properties of Protein-Metal-Organic Frameworks.
J.Am.Chem.Soc., 142, 2020
|
|
2PMF
 
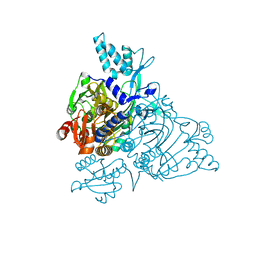 | | The crystal structure of a human glycyl-tRNA synthetase mutant | | Descriptor: | CHLORIDE ION, GLYCEROL, Glycyl-tRNA synthetase | | Authors: | Xie, W. | | Deposit date: | 2007-04-21 | | Release date: | 2007-05-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Long-range structural effects of a Charcot-Marie- Tooth disease-causing mutation in human glycyl-tRNA synthetase.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
7ICP
 
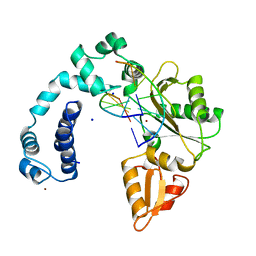 | |
5N5V
 
 | |
