3W5G
 
 | | Crystal structure of tomato beta-galactosidase 4 in complex with galactose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-galactosidase, ... | | Authors: | Eda, M, Tada, T. | | Deposit date: | 2013-01-29 | | Release date: | 2014-01-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of tomato beta-galactosidase 4 in complex with galactose
To be Published
|
|
4ILM
 
 | | CRISPR RNA Processing endoribonuclease | | Descriptor: | CRISPR-associated endoribonuclease Cas6 2, RNA (5'-R(*GP*CP*UP*AP*AP*UP*CP*UP*AP*CP*UP*AP*UP*AP*GP*A)-3') | | Authors: | Shao, Y, Li, H. | | Deposit date: | 2012-12-31 | | Release date: | 2013-03-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.068 Å) | | Cite: | Recognition and cleavage of a nonstructured CRISPR RNA by its processing endoribonuclease Cas6.
Structure, 21, 2013
|
|
4ILL
 
 | |
4ILV
 
 | | Structure of the dioxygenase domain of SACTE_2871, a novel dioxygenase carbohydrate-binding protein fusion from the cellulolytic bacterium Streptomyces sp. SirexAA-E | | Descriptor: | 1,2-ETHANEDIOL, FE (III) ION, Intradiol ring-cleavage dioxygenase | | Authors: | Bianchetti, C.M, Takasuka, T.E, Bergeman, L.F, Harmann, C.H, Fox, B.G. | | Deposit date: | 2013-01-01 | | Release date: | 2013-05-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Fusion of Dioxygenase and Lignin-binding Domains in a Novel Secreted Enzyme from Cellulolytic Streptomyces sp. SirexAA-E.
J.Biol.Chem., 288, 2013
|
|
3W7K
 
 | | Structure of Trypanosoma cruzi dihydroorotate dehydrogenase in complex with MII-6-066 | | Descriptor: | 5-[2-(3,6-dimethoxynaphthalen-2-yl)ethyl]-2,6-dioxo-1,2,3,6-tetrahydropyrimidine-4-carboxylic acid, COBALT HEXAMMINE(III), Dihydroorotate dehydrogenase (fumarate), ... | | Authors: | Inaoka, D.K, Iida, M, Tabuchi, T, Lee, N, Hashimoto, S, Matsuoka, S, Kuranaga, T, Shiba, T, Sakamoto, K, Suzuki, S, Balogun, E.O, Nara, T, Aoki, T, Inoue, M, Honma, T, Tanaka, A, Harada, S, Kita, K. | | Deposit date: | 2013-02-28 | | Release date: | 2014-03-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Structure of Trypanosoma cruzi dihydroorotate dehydrogenase in complex with MII-6-066
To be Published
|
|
3W84
 
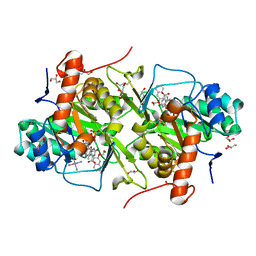 | | Structure of Trypanosoma cruzi dihydroorotate dehydrogenase in complex with MII-6-101 | | Descriptor: | 5-{2-[3-(methoxymethoxy)naphthalen-2-yl]ethyl}-2,6-dioxo-1,2,3,6-tetrahydropyrimidine-4-carboxylic acid, COBALT HEXAMMINE(III), Dihydroorotate dehydrogenase (fumarate), ... | | Authors: | Inaoka, D.K, Iida, M, Tabuchi, T, Lee, N, Hashimoto, S, Matsuoka, S, Kuranaga, T, Shiba, T, Sakamoto, K, Suzuki, S, Balogun, E.O, Nara, T, Aoki, T, Inoue, M, Honma, T, Tanaka, A, Harada, S, Kita, K. | | Deposit date: | 2013-03-12 | | Release date: | 2014-03-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structure of Trypanosoma cruzi dihydroorotate dehydrogenase in complex with MII-6-101
To be Published
|
|
4BBZ
 
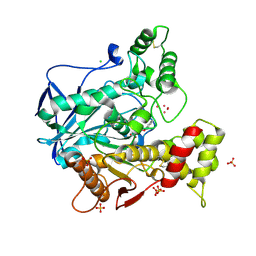 | | Structure of human butyrylcholinesterase inhibited by CBDP (2-min soak): Cresyl-phosphoserine adduct | | Descriptor: | (2-methylphenyl) dihydrogen phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Carletti, E, Colletier, J.-P, Schopfer, L.M, Santoni, G, Masson, P, Lockridge, O, Nachon, F, Weik, M. | | Deposit date: | 2012-09-30 | | Release date: | 2013-02-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Inhibition Pathways of the Potent Organophosphate Cbdp with Cholinesterases Revealed by X-Ray Crystallographic Snapshots and Mass Spectrometry
Chem.Res.Toxicol., 26, 2013
|
|
1W1Y
 
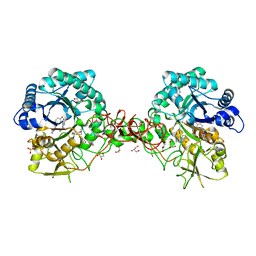 | | Crystal structure of S. marcescens chitinase B in complex with the cyclic dipeptide inhibitor cyclo-(L-Tyr-L-Pro) at 1.85 A resolution | | Descriptor: | CHITINASE B, CYCLO-(L-TYROSINE-L-PROLINE) INHIBITOR, GLYCEROL, ... | | Authors: | Houston, D.R, Synstad, B, Eijsink, V.G.H, Eggleston, I, Van Aalten, D.M.F. | | Deposit date: | 2004-06-24 | | Release date: | 2005-01-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-Based Exploration of Cyclic Dipeptide Chitinase Inhibitors
J.Med.Chem., 47, 2004
|
|
4BJ0
 
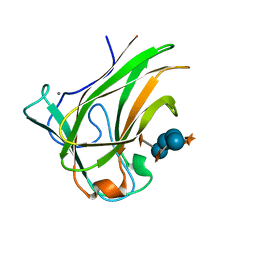 | | Xyloglucan binding module (CBM4-2 X2-L110F) in complex with branched xyloses | | Descriptor: | CALCIUM ION, XYLANASE, alpha-D-glucopyranose, ... | | Authors: | Schantz, L, Hakansson, M, Logan, D.T, Nordberg-Karlsson, E, Ohlin, M. | | Deposit date: | 2013-04-15 | | Release date: | 2014-04-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Carbohydrate Binding Module Recognition of Xyloglucan Defined by Polar Contacts with Branching Xyloses and Ch-Pi Interactions.
Proteins, 82, 2014
|
|
1W08
 
 | | STRUCTURE OF T70N HUMAN LYSOZYME | | Descriptor: | CHLORIDE ION, LYSOZYME | | Authors: | Johnson, R, Christodoulou, J, Luisi, B, Dumoulin, M, Caddy, G, Alcocer, M, Murtagh, G, Archer, D.B, Dobson, C.M. | | Deposit date: | 2004-06-02 | | Release date: | 2004-06-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Rationalising Lysozyme Amyloidosis: Insights from the Structure and Solution Dynamics of T70N Lysozyme.
J.Mol.Biol., 352, 2005
|
|
3WAW
 
 | | Crystal Structure of Autotaxin in Complex with 2BoA | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Nishimasu, H, Ishitani, R, Nureki, O. | | Deposit date: | 2013-05-09 | | Release date: | 2013-07-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.954 Å) | | Cite: | Screening and X-ray Crystal Structure-based Optimization of Autotaxin (ENPP2) Inhibitors, Using a Newly Developed Fluorescence Probe
Acs Chem.Biol., 8, 2013
|
|
2DQD
 
 | | Crystal structure of hyhel-10 FV mutant (Hy50f) complexed with hen egg lysozyme | | Descriptor: | Ig VH,anti-lysozyme, Lysozyme C, lysozyme binding Ig kappa chain V23-J2 region | | Authors: | Shiroishi, M, Kondo, H, Tsumoto, K, Kumagai, I. | | Deposit date: | 2006-05-25 | | Release date: | 2007-01-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural consequences of mutations in interfacial Tyr residues of a protein antigen-antibody complex. The case of HyHEL-10-HEL
J.Biol.Chem., 282, 2007
|
|
1W1T
 
 | | Crystal structure of S. marcescens chitinase B in complex with the cyclic dipeptide inhibitor cyclo-(His-L-Pro) at 1.9 A resolution | | Descriptor: | CHITINASE B, CYCLO-(L-HISTIDINE-L-PROLINE) INHIBITOR, GLYCEROL, ... | | Authors: | Houston, D.R, Synstad, B, Eijsink, V.G.H, Eggleston, I, van Aalten, D.M.F. | | Deposit date: | 2004-06-24 | | Release date: | 2005-01-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-Based Exploration of Cyclic Dipeptide Chitinase Inhibitors
J.Med.Chem., 47, 2004
|
|
4IDA
 
 | |
1W3J
 
 | | Family 1 b-glucosidase from Thermotoga maritima in complex with tetrahydrooxazine | | Descriptor: | BETA-GLUCOSIDASE, TETRAHYDROOXAZINE | | Authors: | Gloster, T.M, Macdonald, J.M, Tarling, C.A, Stick, R.V, Withers, S.W, Davies, G.J. | | Deposit date: | 2004-07-16 | | Release date: | 2004-09-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural, Thermodynamic, and Kinetic Analyses of Tetrahydrooxazine-Derived Inhibitors Bound to {Beta}-Glucosidases
J.Biol.Chem., 279, 2004
|
|
4IDM
 
 | |
2DUU
 
 | | Crystal Structure of apo-form of NADP-Dependent Glyceraldehyde-3-Phosphate Dehydrogenase from Synechococcus Sp. | | Descriptor: | Glyceraldehyde 3-phosphate dehydrogenase, SULFATE ION | | Authors: | Kitatani, T, Nakamura, Y, Wada, K, Kinoshita, T, Tamoi, M, Shigeoka, S, Tada, T. | | Deposit date: | 2006-07-27 | | Release date: | 2006-09-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of apo-glyceraldehyde-3-phosphate dehydrogenase from Synechococcus PCC7942
Acta Crystallogr.,Sect.F, 62, 2006
|
|
4IEY
 
 | | Cys-persulfenate bound Cysteine Dioxygenase at pH 7.0 in the presence of Cys, home-source structure | | Descriptor: | Cysteine dioxygenase type 1, FE (II) ION, S-HYDROPEROXYCYSTEINE | | Authors: | Driggers, C.M, Cooley, R.B, Karplus, P.A. | | Deposit date: | 2012-12-13 | | Release date: | 2013-06-26 | | Last modified: | 2013-09-11 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Cysteine Dioxygenase Structures from pH4 to 9: Consistent Cys-Persulfenate Formation at Intermediate pH and a Cys-Bound Enzyme at Higher pH.
J.Mol.Biol., 425, 2013
|
|
3WDW
 
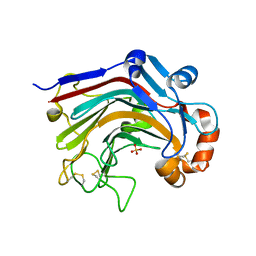 | | The apo-form structure of E113A from Paecilomyces thermophila | | Descriptor: | Beta-1,3-1,4-glucanase, SULFATE ION | | Authors: | Cheng, Y.S, Huang, C.H, Chen, C.C, Huang, T.Y, Ko, T.P, Huang, J.W, Wu, T.H, Liu, J.R, Guo, R.T. | | Deposit date: | 2013-06-25 | | Release date: | 2014-07-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and mutagenetic analyses of a 1,3-1,4-beta-glucanase from Paecilomyces thermophila
Biochim.Biophys.Acta, 1844, 2014
|
|
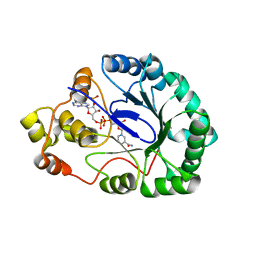
3WBY
 
 | |
3WF4
 
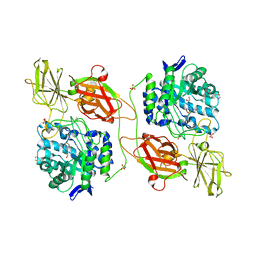 | | Crystal structure of human beta-galactosidase mutant I51T in complex with 6S-NBI-DGJ | | Descriptor: | (3Z,6S,7R,8S,8aS)-3-(butylimino)hexahydro[1,3]thiazolo[3,4-a]pyridine-6,7,8-triol, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Suzuki, H, Ohto, U, Shimizu, T. | | Deposit date: | 2013-07-16 | | Release date: | 2014-04-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of pharmacological chaperoning for human beta-galactosidase
to be published
|
|
1W1V
 
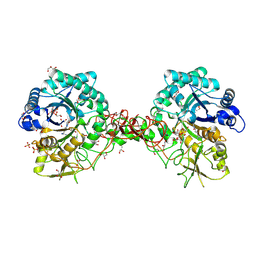 | | Crystal structure of S. marcescens chitinase B in complex with the cyclic dipeptide inhibitor cyclo-(L-Arg-L-Pro) at 1.85 A resolution | | Descriptor: | CHITINASE B, CYCLO-(L-ARGININE-L-PROLINE) INHIBITOR, GLYCEROL, ... | | Authors: | Houston, D.R, Synstad, B, Eijsink, V.G.H, Eggleston, I, Van Aalten, D.M.F. | | Deposit date: | 2004-06-24 | | Release date: | 2005-01-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-Based Exploration of Cyclic Dipeptide Chitinase Inhibitors
J.Med.Chem., 47, 2004
|
|
4BCW
 
 | | Carbonic anhydrase IX mimic in complex with (E)-2-(5-bromo-2- hydroxyphenyl)ethenesulfonic acid | | Descriptor: | (E)-2-(5-BROMO-2-HYDROXYPHENYL)ETHENESULFONIC ACID, CARBONIC ANHYDRASE 2, ZINC ION | | Authors: | Tars, K, Leitans, J, Kazaks, A. | | Deposit date: | 2012-10-03 | | Release date: | 2013-02-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Sulfocoumarins (1,2-Benzoxathiine-2,2-Dioxides): A Class of Potent and Isoform-Selective Inhibitors of Tumor-Associated Carbonic Anhydrases.
J.Med.Chem., 56, 2013
|
|
1W1P
 
 | | Crystal structure of S. marcescens chitinase B in complex with the cyclic dipeptide inhibitor cyclo-(Gly-L-Pro) at 2.1 A resolution | | Descriptor: | CHITINASE B, CYCLO-(GLYCINE-L-PROLINE) INHIBITOR, GLYCEROL, ... | | Authors: | Houston, D.R, Synstad, B, Eijsink, V.G.H, Eggleston, I, Van Aalten, D.M.F. | | Deposit date: | 2004-06-23 | | Release date: | 2005-01-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Based Exploration of Cyclic Dipeptide Chitinase Inhibitors
J.Med.Chem., 47, 2004
|
|
3WJA
 
 | | The crystal structure of human cytosolic NADP(+)-dependent malic enzyme in apo form | | Descriptor: | NADP-dependent malic enzyme | | Authors: | Li, S.-Y, Chen, M.-C, Yang, P.-C, Chan, N.-L, Liu, J.-H, Hung, H.-C. | | Deposit date: | 2013-10-08 | | Release date: | 2014-08-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.548 Å) | | Cite: | Structural characteristics of the nonallosteric human cytosolic malic enzyme.
Biochim.Biophys.Acta, 1844, 2014
|
|
