5T6K
 
 | | Crystal Structure of TgCDPK1 From Toxoplasma Gondii complexed with GW780159X | | Descriptor: | 4-(3-chlorophenyl)-5-(1,5-naphthyridin-2-yl)-1,3-thiazol-2-amine, Calmodulin-domain protein kinase 1, UNKNOWN ATOM OR ION | | Authors: | Jiang, D.Q, Tempel, W, Walker, J.R, El Bakkouri, M, Loppnau, P, Graslund, S, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Hui, R, Lovato, D.V, Osman, K.T, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-09-01 | | Release date: | 2016-11-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of CDPK1 from toxoplasma gondii complexed with SGC-1-19
to be published
|
|
5TDF
 
 | |
5T6Q
 
 | |
5TE2
 
 | | Crystal structure of 7,8-diaminopelargonic acid synthase (BioA) from Mycobacterium tuberculosis, complexed with a mechanism-based inhibitor | | Descriptor: | 1,2-ETHANEDIOL, Adenosylmethionine-8-amino-7-oxononanoate aminotransferase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Maize, K.M, Eiden, C, Aldrich, C.C, Finzel, B.C. | | Deposit date: | 2016-09-20 | | Release date: | 2017-05-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Rational Optimization of Mechanism-Based Inhibitors through Determination of the Microscopic Rate Constants of Inactivation.
J. Am. Chem. Soc., 139, 2017
|
|
5TF6
 
 | | Structure and conformational plasticity of the U6 small nuclear ribonucleoprotein core | | Descriptor: | CHLORIDE ION, GLYCEROL, POTASSIUM ION, ... | | Authors: | Montemayor, E.J, Brow, D.A, Butcher, S.E. | | Deposit date: | 2016-09-24 | | Release date: | 2017-01-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and conformational plasticity of the U6 small nuclear ribonucleoprotein core.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
5THS
 
 | | Crystal Structure of G302A HDAC8 in complex with M344 | | Descriptor: | 1,2-ETHANEDIOL, 4-(dimethylamino)-N-[7-(hydroxyamino)-7-oxoheptyl]benzamide, Histone deacetylase 8, ... | | Authors: | Porter, N.J, Christianson, D.W. | | Deposit date: | 2016-09-30 | | Release date: | 2016-12-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and Functional Influence of the Glycine-Rich Loop G302GGGY on the Catalytic Tyrosine of Histone Deacetylase 8.
Biochemistry, 55, 2016
|
|
5TII
 
 | |
5T9B
 
 | |
5TKI
 
 | |
5TKQ
 
 | | Crystal structure of human 3HAO with zinc bound in the active site | | Descriptor: | 3-hydroxyanthranilate 3,4-dioxygenase, SULFATE ION, ZINC ION | | Authors: | Pidugu, L.S, Toth, E.A. | | Deposit date: | 2016-10-07 | | Release date: | 2017-04-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structures of human 3-hydroxyanthranilate 3,4-dioxygenase with native and non-native metals bound in the active site.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
5TNU
 
 | | S. tokodaii XPB II crystal structure at 3.0 Angstrom resolution | | Descriptor: | CHLORIDE ION, DNA-dependent ATPase XPBII, GLYCEROL, ... | | Authors: | DuPrez, K.T, Hilario, E, Wang, I, Fan, L. | | Deposit date: | 2016-10-14 | | Release date: | 2017-11-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Application of Electrochemical Devices to Characterize the Dynamic Actions of Helicases on DNA.
Anal.Chem., 90, 2018
|
|
5TFZ
 
 | |
5T6G
 
 | | 2.45 A resolution structure of Norovirus 3CL protease in complex with the dipeptidyl inhibitor 7m (hexagonal form) | | Descriptor: | 3-cyclohexyl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-N~2~-(octylsulfonyl)-L-alaninamide, Genome polyprotein | | Authors: | Lovell, S, Battaile, K.P, Mehzabeen, N, Kankanamalage, A.C.G, Kim, Y, Rathnayake, A.D, Damalanka, V.C, Weerawarna, P.M, Doyle, S.T, Alsoudi, A.F, Dissanayake, D.M.P, Chang, K.-O, Groutas, W.C. | | Deposit date: | 2016-09-01 | | Release date: | 2016-11-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure-based exploration and exploitation of the S4 subsite of norovirus 3CL protease in the design of potent and permeable inhibitors.
Eur J Med Chem, 126, 2016
|
|
5TGH
 
 | |
5T7I
 
 | | Crystal structure of galectin-8N in complex with LNnT | | Descriptor: | Galectin-8, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Bohari, M.H, Yu, X, Blanchard, H. | | Deposit date: | 2016-09-05 | | Release date: | 2017-01-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based rationale for differential recognition of lacto- and neolacto- series glycosphingolipids by the N-terminal domain of human galectin-8.
Sci Rep, 6, 2016
|
|
5THU
 
 | | Crystal Structure of G304A HDAC8 in complex with M344 | | Descriptor: | 4-(dimethylamino)-N-[7-(hydroxyamino)-7-oxoheptyl]benzamide, Histone deacetylase 8, POTASSIUM ION, ... | | Authors: | Porter, N.J, Christianson, D.W. | | Deposit date: | 2016-09-30 | | Release date: | 2016-12-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural and Functional Influence of the Glycine-Rich Loop G302GGGY on the Catalytic Tyrosine of Histone Deacetylase 8.
Biochemistry, 55, 2016
|
|
5TCV
 
 | |
5TCY
 
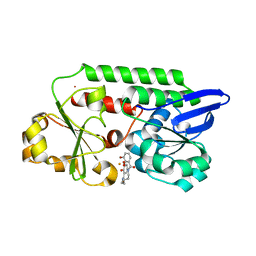 | | A complex of the synthetic siderophore analogue Fe(III)-5-LICAM with CeuE (H227L variant), a periplasmic protein from Campylobacter jejuni. | | Descriptor: | Enterochelin uptake periplasmic binding protein, FE (III) ION, N,N'-pentane-1,5-diylbis(2,3-dihydroxybenzamide) | | Authors: | Wilde, E.J, Blagova, E, Hughes, A, Raines, D.J, Moroz, O.V, Turkenburg, J.P, Duhme-Klair, A.-K, Wilson, K.S. | | Deposit date: | 2016-09-16 | | Release date: | 2017-04-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Interactions of the periplasmic binding protein CeuE with Fe(III) n-LICAM(4-) siderophore analogues of varied linker length.
Sci Rep, 7, 2017
|
|
5T91
 
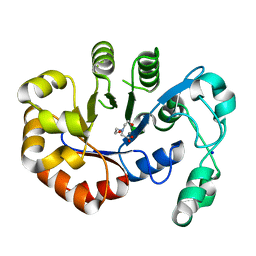 | | Crystal structure of B. subtilis 168 GlpQ in complex with bicine | | Descriptor: | BICINE, CALCIUM ION, Glycerophosphoryl diester phosphodiesterase, ... | | Authors: | Li, F.K.K, Strynadka, N.C.J. | | Deposit date: | 2016-09-09 | | Release date: | 2016-11-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Identification of Two Phosphate Starvation-induced Wall Teichoic Acid Hydrolases Provides First Insights into the Degradative Pathway of a Key Bacterial Cell Wall Component.
J. Biol. Chem., 291, 2016
|
|
5T9T
 
 | | Protocadherin Gamma B2 extracellular cadherin domains 1-5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Protocadherin gamma B2-alpha C, ... | | Authors: | Goodman, K.M, Mannepalli, S, Bahna, F, Honig, B, Shapiro, L. | | Deposit date: | 2016-09-09 | | Release date: | 2016-10-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | gamma-Protocadherin structural diversity and functional implications.
Elife, 5, 2016
|
|
5TDO
 
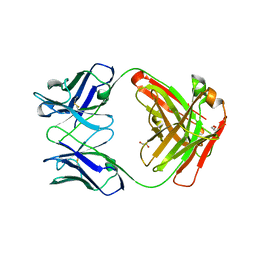 | |
5TE0
 
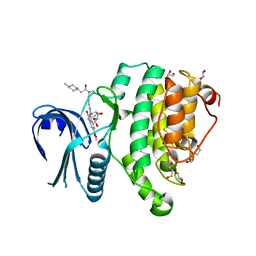 | | Crystal Structure of Adaptor Protein 2 Associated Kinase (AAK1) in complex with BIBF 1120 | | Descriptor: | AP2-associated protein kinase 1, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Counago, R.M, Elkins, J.M, Bountra, C, Arruda, P, Edwards, A.M, Gileadi, O, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-09-20 | | Release date: | 2016-11-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Adaptor Protein 2 Associated Kinase (AAK1) in complex with BIBF 1120
To Be Published
|
|
5TEE
 
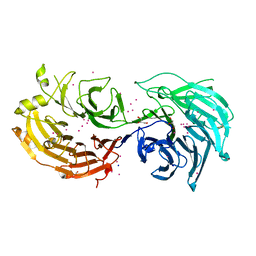 | | Crystal structure of Gemin5 WD40 repeats in apo form | | Descriptor: | GLYCEROL, Gem-associated protein 5, SODIUM ION, ... | | Authors: | Chao, X, Tempel, W, Bian, C, Cerovina, T, He, H, Walker, J.R, Seitova, A, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-09-21 | | Release date: | 2016-10-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural insights into Gemin5-guided selection of pre-snRNAs for snRNP assembly.
Genes Dev., 30, 2016
|
|
5TFO
 
 | |
5TG1
 
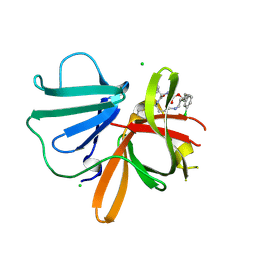 | | 1.40 A resolution structure of Norovirus 3CL protease in complex with the a m-chlorophenyl substituted macrocyclic inhibitor (17-mer) | | Descriptor: | (4S,7S,17S)-17-(3-chlorophenyl)-7-(hydroxymethyl)-4-(2-methylpropyl)-1-oxa-3,6,11-triazacycloheptadecane-2,5,10-trione, 3C-LIKE PROTEASE, CHLORIDE ION | | Authors: | Lovell, S, Battaile, K.P, Mehzabeen, N, Damalanka, V.C, Kim, Y, Kankanamalage, A.C.G, Chang, K.-O, Groutas, W.C. | | Deposit date: | 2016-09-27 | | Release date: | 2017-01-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Design, synthesis, and evaluation of a novel series of macrocyclic inhibitors of norovirus 3CL protease.
Eur J Med Chem, 127, 2016
|
|
