8BCX
 
 | |
8POH
 
 | |
6Y9O
 
 | | Crystal structure of Whirlin PDZ3_C-ter in complex with CASK internal PDZ binding motif peptide | | Descriptor: | Peripheral plasma membrane protein CASK, Whirlin | | Authors: | Zhu, Y, Delhommel, F, Haouz, A, Caillet-Saguy, C, Vaney, M, Mechaly, A.E, Wolff, N. | | Deposit date: | 2020-03-10 | | Release date: | 2020-10-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.632 Å) | | Cite: | Deciphering the Unexpected Binding Capacity of the Third PDZ Domain of Whirlin to Various Cochlear Hair Cell Partners.
J.Mol.Biol., 432, 2020
|
|
8X0S
 
 | | Crystal structure of r(G4C2)2 | | Descriptor: | POTASSIUM ION, RNA (5'-R(*GP*GP*GP*GP*CP*CP*GP*GP*GP*GP*C)-3') | | Authors: | Geng, Y, Liu, C, Cai, Q, Zhu, G. | | Deposit date: | 2023-11-05 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (2.956 Å) | | Cite: | Crystal structure of a tetrameric RNA G-quadruplex formed by hexanucleotide repeat expansions of C9orf72 in ALS/FTD.
Nucleic Acids Res., 52, 2024
|
|
7U1H
 
 | |
7ANY
 
 | | Structure of the Laspartomycin C Friulimicin-like mutant in complex with Geranyl phosphate | | Descriptor: | CADMIUM ION, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Zeronian, M.R, Pearce, N.M, Lutz, M, Wood, T.M, Martin, N.I, Janssen, B.J.C. | | Deposit date: | 2020-10-13 | | Release date: | 2022-02-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.135 Å) | | Cite: | Mechanistic insights into the C55-P targeting lipopeptide antibiotics revealed by structure-activity studies and high-resolution crystal structures
Chem Sci, 13, 2022
|
|
8PNQ
 
 | |
8PNP
 
 | |
5AP2
 
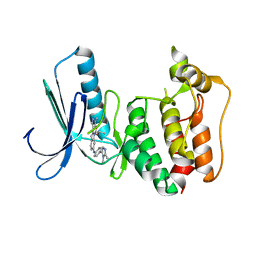 | | Naturally Occurring Mutations in the MPS1 Gene Predispose Cells to Kinase Inhibitor Drug Resistance. | | Descriptor: | DUAL SPECIFICITY PROTEIN KINASE TTK, ISOPROPYL 6-((4-(1,2-DIMETHYL-1H-IMIDAZOL-5-YL)PHENYL)AMINO)-2-(1-METHYL-1H-PYRAZOL-4-YL)-1H-PYRROLO[3,2-C]PYRIDINE-1-CARBOXYLATE | | Authors: | Gurden, M.D, Westwood, I.M, Faisal, A, Naud, S, Cheung, K.J, McAndrew, C, Wood, A, Schmitt, J, Boxall, K, Mak, G, Workman, P, Burke, R, Hoelder, S, Blagg, J, van Montfort, R.L.M, Linardopoulos, S. | | Deposit date: | 2015-09-14 | | Release date: | 2015-09-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Naturally Occurring Mutations in the Mps1 Gene Predispose Cells to Kinase Inhibitor Drug Resistance.
Cancer Res., 75, 2015
|
|
8PM0
 
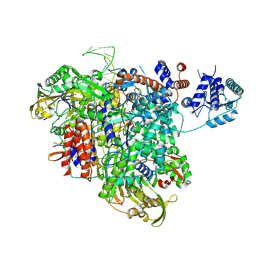 | |
1UTA
 
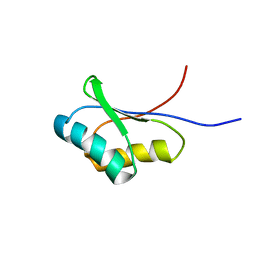 | | Solution structure of the C-terminal RNP domain from the divisome protein FtsN | | Descriptor: | CELL DIVISION PROTEIN FTSN | | Authors: | Yang, J.-C, van den Ent, F, Neuhaus, D, Brevier, J, Lowe, J. | | Deposit date: | 2003-12-04 | | Release date: | 2004-09-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure and Domain Architecture of the Divisome Protein Ftsn
Mol.Microbiol., 52, 2004
|
|
5HHI
 
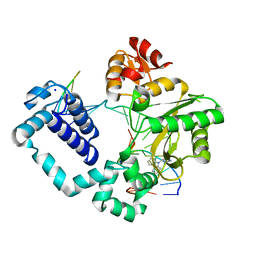 | | Structure of human DNA polymerase beta Host-Guest complexed with CBZ-platinated N7-G | | Descriptor: | DNA (5'-D(*CP*CP*GP*AP*CP*GP*GP*AP*GP*GP*AP*GP*CP*AP*GP*G)-3'), DNA (5'-D(P*CP*CP*TP*GP*CP*TP*CP*CP*TP*C)-3'), DNA (5'-D(P*GP*TP*CP*GP*G)-3'), ... | | Authors: | Lee, S, Koag, M.-C. | | Deposit date: | 2016-01-11 | | Release date: | 2017-01-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.517 Å) | | Cite: | Synthesis, structure, and biological evaluation of a platinum-carbazole conjugate.
Chem Biol Drug Des, 91, 2018
|
|
6PIV
 
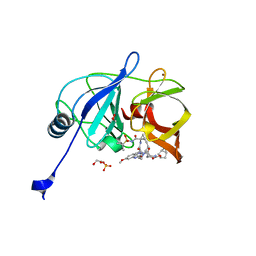 | | Crystal structure of HCV NS3/4A D168A protease in complex with P4-7 (NR03-77) | | Descriptor: | (1R,3r,5S)-bicyclo[3.1.0]hexan-3-yl [(2R,6S,12Z,13aS,14aR,16aS)-2-[(7-methoxy-3-methylquinoxalin-2-yl)oxy]-14a-{[(1-methylcyclopropyl)sulfonyl]carbamoyl}-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl]carbamate, 1,2-ETHANEDIOL, NS3/4A protease, ... | | Authors: | Zephyr, J, Schiffer, C.A. | | Deposit date: | 2019-06-27 | | Release date: | 2020-03-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Avoiding Drug Resistance by Substrate Envelope-Guided Design: Toward Potent and Robust HCV NS3/4A Protease Inhibitors.
Mbio, 11, 2020
|
|
6PJ0
 
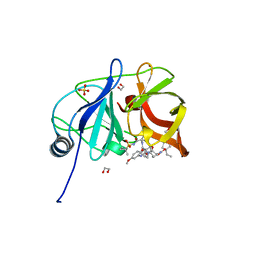 | | Crystal structure of HCV NS3/4A D168A protease in complex with P4-5 (NR01-97) | | Descriptor: | 1,2-ETHANEDIOL, 1-ethylcyclopentyl [(2R,6S,12Z,13aS,14aR,16aS)-2-[(7-methoxy-3-methylquinoxalin-2-yl)oxy]-14a-{[(1-methylcyclopropyl)sulfonyl]carbamoyl}-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl]carbamate, NS3/4 Aprotease, ... | | Authors: | Zephyr, J, Schiffer, C.A. | | Deposit date: | 2019-06-27 | | Release date: | 2020-03-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Avoiding Drug Resistance by Substrate Envelope-Guided Design: Toward Potent and Robust HCV NS3/4A Protease Inhibitors.
Mbio, 11, 2020
|
|
6MAV
 
 | |
8IED
 
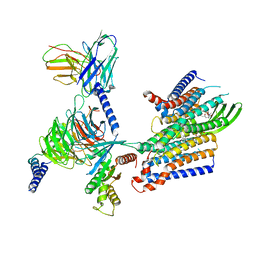 | | Cryo-EM structure of GPR156-miniGo-scFv16 complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(o) subunit alpha, ... | | Authors: | Shin, J, Park, J, Cho, Y. | | Deposit date: | 2023-02-15 | | Release date: | 2024-02-14 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Constitutive activation mechanism of a class C GPCR.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8IEP
 
 | |
5HHH
 
 | | Structure of human DNA polymerase beta Host-Guest complexed with the control G for N7-CBZ-platination | | Descriptor: | DNA (5'-D(*CP*CP*GP*AP*CP*GP*GP*AP*GP*GP*AP*GP*CP*AP*GP*G)-3'), DNA (5'-D(P*CP*CP*TP*GP*CP*TP*CP*CP*TP*C)-3'), DNA (5'-D(P*GP*TP*CP*GP*G)-3'), ... | | Authors: | Koag, M.-C, Lee, S. | | Deposit date: | 2016-01-11 | | Release date: | 2017-01-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.363 Å) | | Cite: | Synthesis, structure, and biological evaluation of a platinum-carbazole conjugate.
Chem Biol Drug Des, 91, 2018
|
|
6PIW
 
 | |
6PJ1
 
 | | Crystal structure of HCV NS3/4A D168A protease in complex with P4-4(AJ-74) | | Descriptor: | 1,2-ETHANEDIOL, 1-methylcyclopentyl [(2R,6S,12Z,13aS,14aR,16aS)-2-[(7-methoxy-3-methylquinoxalin-2-yl)oxy]-14a-{[(1-methylcyclopropyl)sulfonyl]carbamoyl}-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl]carbamate, NS3/4A protease, ... | | Authors: | Zephyr, J, Schiffer, C.A. | | Deposit date: | 2019-06-27 | | Release date: | 2020-03-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Avoiding Drug Resistance by Substrate Envelope-Guided Design: Toward Potent and Robust HCV NS3/4A Protease Inhibitors.
Mbio, 11, 2020
|
|
8IEQ
 
 | | Cryo-EM structure of G-protein free GPR156 | | Descriptor: | Probable G-protein coupled receptor 156, [(2R)-3-[(E)-hexadec-9-enoyl]oxy-2-octadecanoyloxy-propyl] 2-(trimethylazaniumyl)ethyl phosphate | | Authors: | Shin, J, Park, J, Cho, Y. | | Deposit date: | 2023-02-15 | | Release date: | 2024-02-14 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | Constitutive activation mechanism of a class C GPCR.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8B1P
 
 | | Crystal structure of SUDV VP40 CCS mutant | | Descriptor: | Matrix protein VP40 | | Authors: | Werner, A.-D, Becker, S. | | Deposit date: | 2022-09-11 | | Release date: | 2023-06-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The C-terminus of Sudan ebolavirus VP40 contains a functionally important CX n C motif, a target for redox modifications.
Structure, 31, 2023
|
|
8B1O
 
 | | Crystal structure of SUDV VP40 C314S mutant | | Descriptor: | Matrix protein VP40 | | Authors: | Werner, A.-D, Becker, S. | | Deposit date: | 2022-09-11 | | Release date: | 2023-06-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The C-terminus of Sudan ebolavirus VP40 contains a functionally important CX n C motif, a target for redox modifications.
Structure, 31, 2023
|
|
7KP6
 
 | | Structure of Ack1 kinase in complex with a selective inhibitor | | Descriptor: | 5-chloro-N~2~-[4-(4-methylpiperazin-1-yl)phenyl]-N~4~-{[(2R)-oxolan-2-yl]methyl}pyrimidine-2,4-diamine, Activated CDC42 kinase 1, CHLORIDE ION | | Authors: | Thakur, M.K, Miller, W.T, Mahajan, N, Seeliger, M.A. | | Deposit date: | 2020-11-10 | | Release date: | 2022-02-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Inhibiting ACK1-mediated phosphorylation of C-terminal Src kinase counteracts prostate cancer immune checkpoint blockade resistance.
Nat Commun, 13, 2022
|
|
9FRE
 
 | | CryoEM structure of human rho1 GABAA receptor in complex with THIP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4,5,6,7-tetrahydro-[1,2]oxazolo[5,4-c]pyridin-3-one, CHLORIDE ION, ... | | Authors: | Fan, C, Howard, R.J, Lindahl, E. | | Deposit date: | 2024-06-18 | | Release date: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.19 Å) | | Cite: | Cryo-EM structures of Rho1 GABAA receptors with antagonist and agonist drugs
To Be Published
|
|
