2Q5Z
 
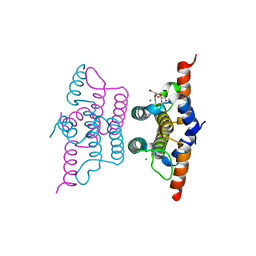 | | Crystal structure of iMazG from Vibrio DAT 722: Ntag-iMazG (P43212) | | Descriptor: | GLYCEROL, Hypothetical protein, MAGNESIUM ION | | Authors: | Robinson, A, Guilfoyle, A.P, Harrop, S.J, Boucher, Y, Stokes, H.W, Curmi, P.M.G, Mabbutt, B.C. | | Deposit date: | 2007-06-04 | | Release date: | 2007-10-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A putative house-cleaning enzyme encoded within an integron array: 1.8 A crystal structure defines a new MazG subtype.
Mol.Microbiol., 66, 2007
|
|
3VAV
 
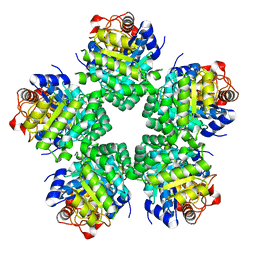 | |
2PTY
 
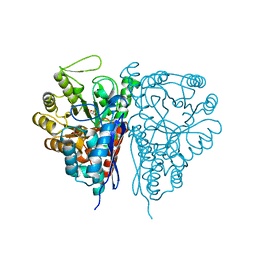 | | Crystal Structure of the T. brucei enolase complexed with PEP | | Descriptor: | 1,2-ETHANEDIOL, Enolase, PHOSPHOENOLPYRUVATE, ... | | Authors: | Navarro, M.V.A.S, Rigden, D.J, Garratt, R.C, Dias, S.M.G. | | Deposit date: | 2007-05-08 | | Release date: | 2007-11-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural flexibility in Trypanosoma brucei enolase revealed by X-ray crystallography and molecular dynamics.
Febs J., 274, 2007
|
|
6IBM
 
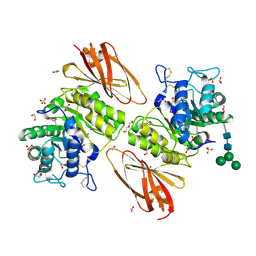 | | Crystal structure of human alpha-galactosidase A in complex with alpha-galactose configured cyclosulfate ME776 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Rowland, R.J, Wu, L, Davies, G.J. | | Deposit date: | 2018-11-30 | | Release date: | 2019-10-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Alpha-d-Gal-cyclophellitol cyclosulfamidate is a Michaelis complex analog that stabilizes therapeutic lysosomal alpha-galactosidase A in Fabry disease
Chem Sci, 2019
|
|
3QWM
 
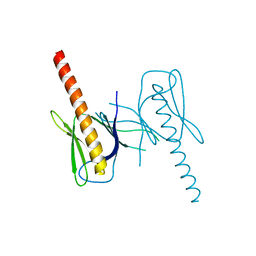 | | Crystal Structure of GEP100, the plextrin homology domain of IQ motif and SEC7 domain-containing protein 1 isoform a | | Descriptor: | IQ motif and SEC7 domain-containing protein 1 | | Authors: | Filippakopoulos, P, Picaud, S, Felletar, I, Feller, S, Janning, M, Sabe, H, Krojer, T, Chaikuad, A, Allerston, C, von Delft, F, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-02-28 | | Release date: | 2011-04-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Crystal Structure of GEP100, the plextrin homology domain of IQ motif and SEC7 domain-containing protein 1 isoform a
to be published
|
|
3VWD
 
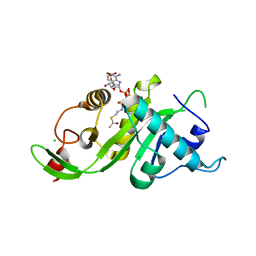 | |
7BEN
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 Spike glycoprotein in a ternary complex with COVOX-253 and COVOX-75 Fabs | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, BROMIDE ION, ... | | Authors: | Zhou, D, Zhao, Y, Ren, J, Stuart, D. | | Deposit date: | 2020-12-24 | | Release date: | 2021-03-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The antigenic anatomy of SARS-CoV-2 receptor binding domain.
Cell, 184, 2021
|
|
3QVC
 
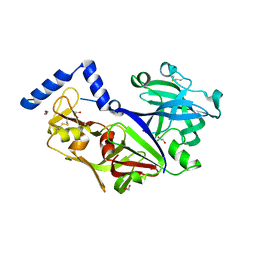 | |
4O76
 
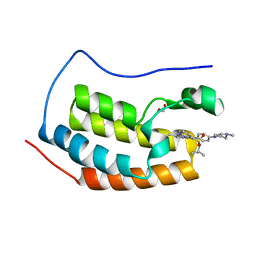 | | Crystal structure of the first bromodomain of human BRD4 in complex with TG101209 | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, N-tert-butyl-3-[(5-methyl-2-{[4-(4-methylpiperazin-1-yl)phenyl]amino}pyrimidin-4-yl)amino]benzenesulfonamide | | Authors: | Zhu, J.-Y, Ember, S.W, Watts, C, Schonbrunn, E. | | Deposit date: | 2013-12-24 | | Release date: | 2014-03-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Acetyl-lysine Binding Site of Bromodomain-Containing Protein 4 (BRD4) Interacts with Diverse Kinase Inhibitors.
Acs Chem.Biol., 9, 2014
|
|
5MZV
 
 | | IL-23:IL-23R:Nb22E11 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Bloch, Y, Savvides, S.N. | | Deposit date: | 2017-02-01 | | Release date: | 2018-01-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Activation of Pro-inflammatory Human Cytokine IL-23 by Cognate IL-23 Receptor Enables Recruitment of the Shared Receptor IL-12R beta 1.
Immunity, 48, 2018
|
|
1N4L
 
 | | A DNA analogue of the polypurine tract of HIV-1 | | Descriptor: | 5'-D(*CP*TP*TP*TP*TP*CP*TP*TP*TP*TP*AP*AP*AP*AP*AP*G)-3', 5'-D(*CP*TP*TP*TP*TP*TP*AP*AP*AP*AP*GP*AP*AP*AP*AP*G)-3', Reverse Transcriptase | | Authors: | Cote, M.L, Pflomm, M, Georgiadis, M.M. | | Deposit date: | 2002-10-31 | | Release date: | 2003-06-24 | | Last modified: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Staying Straight with A-tracts: A DNA Analog of the HIV-1 Polypurine Tract
J.Mol.Biol., 330, 2003
|
|
3VAF
 
 | | Structure of U2AF65 variant with BrU3 DNA | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, DNA 5'-D(*UP*UP*(BRU)P*(BRU)P*UP*UP*U)-3', GLYCEROL, ... | | Authors: | Jenkins, J.L, Kielkopf, C.L. | | Deposit date: | 2011-12-29 | | Release date: | 2013-02-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | U2AF65 adapts to diverse pre-mRNA splice sites through conformational selection of specific and promiscuous RNA recognition motifs.
Nucleic Acids Res., 41, 2013
|
|
3VAM
 
 | | Structure of U2AF65 variant with BrU5C2 DNA | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, DNA (5'-D(*UP*CP*UP*UP*(BRU)P*UP*U)-3'), GLYCEROL, ... | | Authors: | Jenkins, J.L, Kielkopf, C.L. | | Deposit date: | 2011-12-29 | | Release date: | 2013-02-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | U2AF65 adapts to diverse pre-mRNA splice sites through conformational selection of specific and promiscuous RNA recognition motifs.
Nucleic Acids Res., 41, 2013
|
|
3FJB
 
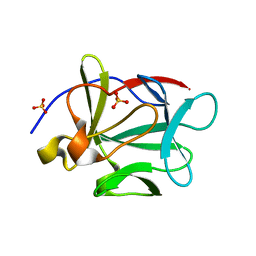 | |
1BAF
 
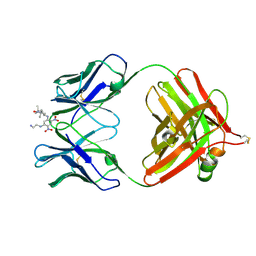 | | 2.9 ANGSTROMS RESOLUTION STRUCTURE OF AN ANTI-DINITROPHENYL-SPIN-LABEL MONOCLONAL ANTIBODY FAB FRAGMENT WITH BOUND HAPTEN | | Descriptor: | IGG1-KAPPA AN02 FAB (HEAVY CHAIN), IGG1-KAPPA AN02 FAB (LIGHT CHAIN), N-(2-AMINO-ETHYL)-4,6-DINITRO-N'-(2,2,6,6-TETRAMETHYL-1-OXY-PIPERIDIN-4-YL)-BENZENE-1,3-DIAMINE | | Authors: | Leahy, D.J, Brunger, A.T, Fox, R.O, Hynes, T.R. | | Deposit date: | 1992-01-16 | | Release date: | 1994-01-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | 2.9 A resolution structure of an anti-dinitrophenyl-spin-label monoclonal antibody Fab fragment with bound hapten.
J.Mol.Biol., 221, 1991
|
|
4JZE
 
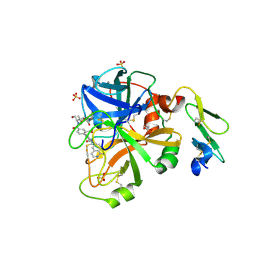 | | Structure of factor VIIA in complex with the inhibitor 2-{2-[(1-aminoisoquinolin-6-yl)carbamoyl]-6-methoxypyridin-3-yl}-5-{[(2S)-1-hydroxy-3,3-dimethylbutan-2-yl]carbamoyl}benzoic acid | | Descriptor: | 2-{2-[(1-aminoisoquinolin-6-yl)carbamoyl]-6-methoxypyridin-3-yl}-5-{[(2S)-1-hydroxy-3,3-dimethylbutan-2-yl]carbamoyl}benzoic acid, CALCIUM ION, Factor VIIa (Heavy Chain), ... | | Authors: | Wei, A, Anumula, R. | | Deposit date: | 2013-04-02 | | Release date: | 2013-08-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Discovery of nonbenzamidine factor VIIa inhibitors using a biaryl acid scaffold.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
3R7R
 
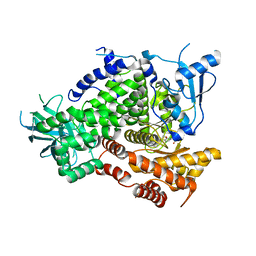 | | Structure-based design of thienobenzoxepin inhibitors of PI3-Kinase | | Descriptor: | 8-(acetylamino)-N-(2-chlorophenyl)-N-methyl-4,5-dihydrothieno[3,2-d][1]benzoxepine-2-carboxamide, Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma isoform | | Authors: | Murray, J.M, Wiesmann, C. | | Deposit date: | 2011-03-22 | | Release date: | 2011-08-03 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure-based design of thienobenzoxepin inhibitors of PI3-kinase.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
2A15
 
 | | X-ray Crystal Structure of RV0760 from Mycobacterium Tuberculosis at 1.68 Angstrom Resolution | | Descriptor: | HYPOTHETICAL PROTEIN Rv0760c, NICOTINAMIDE | | Authors: | Garen, C.R, Cherney, M.M, James, M.N.G, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2005-06-17 | | Release date: | 2005-10-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Crystal structure of Mycobacterium tuberculosis Rv0760c at 1.50 A resolution, a structural homolog of Delta(5)-3-ketosteroid isomerase
Biochim.Biophys.Acta, 1784, 2008
|
|
5QOK
 
 | | PanDDA analysis group deposition -- Crystal Structure of DCP2 (NUDT20) in complex with FMOPL000294a | | Descriptor: | 1,2-ETHANEDIOL, 8-[(dimethylamino)methyl]-4-methyl-7-oxidanyl-chromen-2-one, ACETATE ION, ... | | Authors: | Nelson, E.R, Velupillai, S, Talon, R, Collins, P.M, Krojer, T, Wang, D, Brandao-Neto, J, Douangamath, A, Burgess-Brown, N, Arrowsmith, C.H, Bountra, C, Huber, K, von Delft, F. | | Deposit date: | 2019-02-22 | | Release date: | 2019-05-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
3RAQ
 
 | |
4FI8
 
 | | Kinetic Stabilization of transthyretin through covalent modification of K15 by 4-bromo-3-(5-(3,5-dichloro-4-hydroxyphenyl)-1,3,4-oxadiazol-2-yl)-benzenesulfonamide | | Descriptor: | 4-bromo-3-[5-(3,5-dichloro-4-hydroxyphenyl)-1,3,4-oxadiazol-2-yl]benzenesulfonyl fluoride, Transthyretin | | Authors: | Connelly, S, Grimster, N, Wilson, I.A, Kelly, J.W. | | Deposit date: | 2012-06-08 | | Release date: | 2013-02-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Aromatic Sulfonyl Fluorides Covalently Kinetically Stabilize Transthyretin to Prevent Amyloidogenesis while Affording a Fluorescent Conjugate.
J.Am.Chem.Soc., 135, 2013
|
|
4AW5
 
 | | Complex of the EphB4 kinase domain with an oxindole inhibitor | | Descriptor: | (3Z)-5-[(1-ethylpiperidin-4-yl)amino]-3-[(5-methoxy-1H-benzimidazol-2-yl)(phenyl)methylidene]-1,3-dihydro-2H-indol-2-one, EPHRIN TYPE-B RECEPTOR 4 | | Authors: | Till, J.H, Stout, T.J. | | Deposit date: | 2012-05-31 | | Release date: | 2012-08-01 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | The Design, Synthesis, and Biological Evaluation of Potent Receptor Tyrosine Kinase Inhibitors.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
7BD7
 
 | |
257L
 
 | | AN ADAPTABLE METAL-BINDING SITE ENGINEERED INTO T4 LYSOZYME | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, PROTEIN (LYSOZYME) | | Authors: | Wray, J.W, Baase, W.A, Ostheimer, G.J, Matthews, B.W. | | Deposit date: | 1999-01-05 | | Release date: | 2000-09-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Use of a non-rigid region in T4 lysozyme to design an adaptable metal-binding site.
Protein Eng., 13, 2000
|
|
4K7U
 
 | | Crystal structure of Zn2.3-hUb (human ubiquitin) adduct from a solution 70 mM zinc acetate/1.3 mM hUb | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Fermani, S, Falini, G, Calvaresi, M, Bottoni, A, Arnesano, F, Natile, G. | | Deposit date: | 2013-04-17 | | Release date: | 2013-05-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Conformational selection of ubiquitin quaternary structures driven by zinc ions.
Chemistry, 19, 2013
|
|
