6WMQ
 
 | |
5FKK
 
 | | TetR(D) N82A mutant in complex with anhydrotetracycline and magnesium | | Descriptor: | 5A,6-ANHYDROTETRACYCLINE, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Werten, S, Schneider, J, Palm, G.J, Hinrichs, W. | | Deposit date: | 2015-10-17 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Modular Organisation of Inducer Recognition and Allostery in the Tetracycline Repressor
FEBS J., 283, 2016
|
|
5FKM
 
 | | TetR(D) T103A mutant in complex with anhydrotetracycline and magnesium, I4(1)22 | | Descriptor: | 5A,6-ANHYDROTETRACYCLINE, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Werten, S, Schneider, J, Palm, G.J, Hinrichs, W. | | Deposit date: | 2015-10-17 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Modular Organisation of Inducer Recognition and Allostery in the Tetracycline Repressor
FEBS J., 283, 2016
|
|
5FKN
 
 | | TetR(D) T103A mutant in complex with anhydrotetracycline and magnesium, P4(3)2(1)2 | | Descriptor: | 5A,6-ANHYDROTETRACYCLINE, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Werten, S, Schneider, J, Palm, G.J, Hinrichs, W. | | Deposit date: | 2015-10-17 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Modular Organisation of Inducer Recognition and Allostery in the Tetracycline Repressor
FEBS J., 283, 2016
|
|
5FKL
 
 | | TetR(D) H100A mutant in complex with anhydrotetracycline and magnesium | | Descriptor: | 5A,6-ANHYDROTETRACYCLINE, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Werten, S, Schneider, J, Palm, G.J, Hinrichs, W. | | Deposit date: | 2015-10-17 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Modular Organisation of Inducer Recognition and Allostery in the Tetracycline Repressor
FEBS J., 283, 2016
|
|
7KFO
 
 | | Crystal structure of the MarR family transcriptional regulator from Variovorax paradoxus bound to Indole 3 acetic acid | | Descriptor: | 1H-INDOL-3-YLACETIC ACID, SULFATE ION, Transcriptional regulator, ... | | Authors: | Walton, W.G, Redinbo, M.R, Dangl, J.L. | | Deposit date: | 2020-10-14 | | Release date: | 2021-11-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Diverse MarR bacterial regulators of auxin catabolism in the plant microbiome.
Nat Microbiol, 7, 2022
|
|
7KUA
 
 | | Crystal structure of the MarR family transcriptional regulator from Pseudomonas putida bound to Indole 3 acetic acid | | Descriptor: | 1H-INDOL-3-YLACETIC ACID, Transcriptional regulator, MarR family | | Authors: | Walton, W.G, Lietzan, A.D, Redinbo, M.R, Dangl, J.L. | | Deposit date: | 2020-11-24 | | Release date: | 2021-12-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Diverse MarR bacterial regulators of auxin catabolism in the plant microbiome.
Nat Microbiol, 7, 2022
|
|
7L19
 
 | | Crystal structure of the MarR family transcriptional regulator from Enterobacter soli strain LF7 bound to Indole 3 acetic acid | | Descriptor: | 1H-INDOL-3-YLACETIC ACID, MarR family transcriptional regulator, NICKEL (II) ION | | Authors: | Lietzan, A.D, Walton, W.G, Redinbo, M.R, Dangl, J.L. | | Deposit date: | 2020-12-14 | | Release date: | 2021-12-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Diverse MarR bacterial regulators of auxin catabolism in the plant microbiome.
Nat Microbiol, 7, 2022
|
|
7L1I
 
 | | Crystal structure of the MarR family transcriptional regulator from Acineotobacter baumannii bound to Indole 3 acetic acid | | Descriptor: | 1H-INDOL-3-YLACETIC ACID, MarR family multidrug resistance pump transcriptional regulator, NICKEL (II) ION | | Authors: | Walton, W.G, Lietzan, A.D, Redinbo, M.R, Dangl, J.L. | | Deposit date: | 2020-12-14 | | Release date: | 2022-02-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Diverse MarR bacterial regulators of auxin catabolism in the plant microbiome.
Nat Microbiol, 7, 2022
|
|
6H49
 
 | | A polyamorous repressor: deciphering the evolutionary strategy used by the phage-inducible chromosomal islands to spread in nature. | | Descriptor: | Orf20, SULFATE ION | | Authors: | Ciges-Tomas, J.R, Alite, C, Bowring, J.Z, Donderis, J, Penades, J.R, Marina, A. | | Deposit date: | 2018-07-20 | | Release date: | 2019-08-28 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structure of a polygamous repressor reveals how phage-inducible chromosomal islands spread in nature.
Nat Commun, 10, 2019
|
|
1G3S
 
 | | CYS102SER DTXR | | Descriptor: | DIPHTHERIA TOXIN REPRESSOR | | Authors: | Pohl, E, Goranson-Siekierke, J, Holmes, R.K, Hol, W.G.J. | | Deposit date: | 2000-10-25 | | Release date: | 2001-10-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of three diphtheria toxin repressor (DtxR) variants with decreased repressor activity.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1G3Y
 
 | | ARG80ALA DTXR | | Descriptor: | DIPHTHERIA TOXIN REPRESSOR, ZINC ION | | Authors: | Pohl, E, Goranson-Siekierke, J, Holmes, R.K, Hol, W.G.J. | | Deposit date: | 2000-10-25 | | Release date: | 2001-10-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of three diphtheria toxin repressor (DtxR) variants with decreased repressor activity.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1G3T
 
 | | CYS102SER DTXR | | Descriptor: | DIPHTHERIA TOXIN REPRESSOR | | Authors: | Pohl, E, Goranson-Siekierke, J, Holmes, R.K, Hol, W.G.J. | | Deposit date: | 2000-10-25 | | Release date: | 2001-10-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structures of three diphtheria toxin repressor (DtxR) variants with decreased repressor activity.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
6C28
 
 | | Transcriptional repressor, CouR, bound to p-coumaroyl-CoA | | Descriptor: | Transcriptional regulator, MarR family, p-coumaroyl-CoA | | Authors: | Cogan, D.P, Nair, S.K. | | Deposit date: | 2018-01-07 | | Release date: | 2018-05-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structural basis of transcriptional regulation by CouR, a repressor of coumarate catabolism, inRhodopseudomonas palustris.
J. Biol. Chem., 293, 2018
|
|
6C9T
 
 | | Transcriptional repressor, CouR | | Descriptor: | CouR | | Authors: | Cogan, D.P, Nair, S.K. | | Deposit date: | 2018-01-28 | | Release date: | 2018-05-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structural basis of transcriptional regulation by CouR, a repressor of coumarate catabolism, inRhodopseudomonas palustris.
J. Biol. Chem., 293, 2018
|
|
3IL2
 
 | |
1G3W
 
 | | CD-CYS102SER DTXR | | Descriptor: | CADMIUM ION, DIPHTHERIA TOXIN REPRESSOR, SULFATE ION | | Authors: | Pohl, E, Goranson-Siekierke, J, Holmes, R.K, Hol, W.G.J. | | Deposit date: | 2000-10-25 | | Release date: | 2001-10-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of three diphtheria toxin repressor (DtxR) variants with decreased repressor activity.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
3IKV
 
 | |
3IKT
 
 | |
3FK6
 
 | | Crystal structure of TetR triple mutant (H64K, S135L, S138I) | | Descriptor: | Tetracycline repressor protein class B from transposon Tn10, Tetracycline repressor protein class D | | Authors: | Klieber, M.A, Scholz, O, Lochner, S, Gmeiner, P, Hillen, W, Muller, Y.A. | | Deposit date: | 2008-12-16 | | Release date: | 2009-10-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural origins for selectivity and specificity in an engineered bacterial repressor-inducer pair.
Febs J., 276, 2009
|
|
6DKS
 
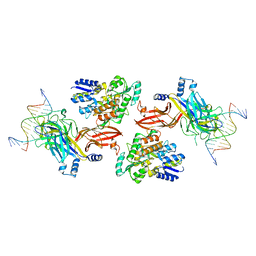 | | Structure of the Rbpj-SHARP-DNA Repressor Complex | | Descriptor: | DNA (5'-D(*AP*AP*TP*CP*TP*TP*TP*CP*CP*CP*AP*CP*AP*GP*T)-3'), DNA (5'-D(*TP*TP*AP*CP*TP*GP*TP*GP*GP*GP*AP*AP*AP*GP*A)-3'), Maltose/maltodextrin-binding periplasmic protein, ... | | Authors: | Kovall, R.A, VanderWielen, B.D, Yuan, Z. | | Deposit date: | 2018-05-30 | | Release date: | 2019-01-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Structural and Functional Studies of the RBPJ-SHARP Complex Reveal a Conserved Corepressor Binding Site.
Cell Rep, 26, 2019
|
|
6SY6
 
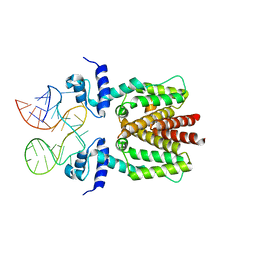 | | TetR in complex with the TetR-binding RNA-aptamer K2 | | Descriptor: | RNA (36-MER), Tetracycline repressor protein class B from transposon Tn10 | | Authors: | Grau, F.C, Muller, Y.A, Suess, B, Groher, F, Jaeger, J. | | Deposit date: | 2019-09-27 | | Release date: | 2020-02-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The complex formed between a synthetic RNA aptamer and the transcription repressor TetR is a structural and functional twin of the operator DNA-TetR regulator complex.
Nucleic Acids Res., 48, 2020
|
|
6SY4
 
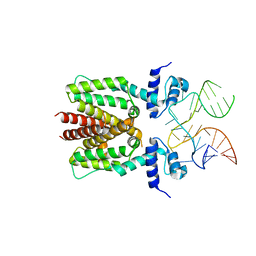 | | TetR in complex with the TetR-binding RNA-aptamer K1 | | Descriptor: | TetR-binding aptamer K1 (43-MER), Tetracycline repressor protein class B from transposon Tn10 | | Authors: | Grau, F.C, Muller, Y.A, Suess, B, Groher, F, Jaeger, J. | | Deposit date: | 2019-09-27 | | Release date: | 2020-02-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.695 Å) | | Cite: | The complex formed between a synthetic RNA aptamer and the transcription repressor TetR is a structural and functional twin of the operator DNA-TetR regulator complex.
Nucleic Acids Res., 48, 2020
|
|
6H4B
 
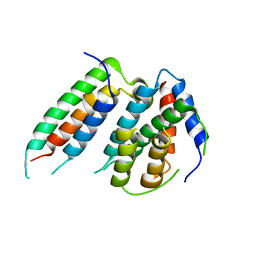 | | A polyamorous repressor: deciphering the evolutionary strategy used by the phage-inducible chromosomal islands to spread in nature. | | Descriptor: | ORF026, Orf20 | | Authors: | Ciges-Tomas, J.R, Alite, C, Bowring, J.Z, Donderis, J, Penades, J.R, Marina, A. | | Deposit date: | 2018-07-20 | | Release date: | 2019-08-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.902 Å) | | Cite: | The structure of a polygamous repressor reveals how phage-inducible chromosomal islands spread in nature.
Nat Commun, 10, 2019
|
|
2XPU
 
 | | TetR(D) in complex with anhydrotetracycline. | | Descriptor: | 5A,6-ANHYDROTETRACYCLINE, CHLORIDE ION, SULFATE ION, ... | | Authors: | Dalm, D, Palm, G.J, Hinrichs, W. | | Deposit date: | 2010-08-30 | | Release date: | 2011-09-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Tetracycline Repressor Allostery Does not Depend on Divalent Metal Recognition.
Biochemistry, 53, 2014
|
|
