6ZHZ
 
 | | OleP-oleandolide(DEO) in high salt crystallization conditions | | Descriptor: | (3~{R},4~{S},5~{R},6~{S},7~{S},9~{S},11~{R},12~{S},13~{R},14~{R})-3,5,7,9,11,13,14-heptamethyl-4,6,12-tris(oxidanyl)-1-oxacyclotetradecane-2,10-dione, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Cytochrome P-450, ... | | Authors: | Montemiglio, L.C, Savino, C, Vallone, B, Parisi, G, Cecchetti, C. | | Deposit date: | 2020-06-24 | | Release date: | 2020-10-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Dissecting the Cytochrome P450 OleP Substrate Specificity: Evidence for a Preferential Substrate.
Biomolecules, 10, 2020
|
|
2J76
 
 | | Solution structure and RNA interactions of the RNA recognition motif from eukaryotic translation initiation factor 4B | | Descriptor: | EUKARYOTIC TRANSLATION INITIATION FACTOR 4B | | Authors: | Fleming, K, Ghuman, J, Yuan, X.M, Simpson, P, Szendroi, A, Matthews, S, Curry, S. | | Deposit date: | 2006-10-06 | | Release date: | 2008-10-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure and RNA Interactions of the RNA Recognition Motif from Eukaryotic Translation Initiation Factor 4B.
Biochemistry, 42, 2003
|
|
5EQH
 
 | | Human GLUT1 in complex with inhibitor (2~{S})-3-(2-bromophenyl)-2-[2-(4-methoxyphenyl)ethanoylamino]-~{N}-[(1~{S})-1-phenylethyl]propanamide | | Descriptor: | (2~{S})-3-(2-bromophenyl)-2-[2-(4-methoxyphenyl)ethanoylamino]-~{N}-[(1~{S})-1-phenylethyl]propanamide, Solute carrier family 2, facilitated glucose transporter member 1 | | Authors: | Kapoor, K, Finer-Moore, J, Pedersen, B.P, Caboni, L, Waight, A.B, Hillig, R, Bringmann, P, Heisler, I, Muller, T, Siebeneicher, H, Stroud, R.M. | | Deposit date: | 2015-11-12 | | Release date: | 2016-04-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Mechanism of inhibition of human glucose transporter GLUT1 is conserved between cytochalasin B and phenylalanine amides.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
3EUL
 
 | | Structure of the signal receiver domain of the putative response regulator NarL from Mycobacterium tuberculosis | | Descriptor: | CHLORIDE ION, POSSIBLE NITRATE/NITRITE RESPONSE TRANSCRIPTIONAL REGULATORY PROTEIN NARL (DNA-binding response regulator, LuxR family) | | Authors: | Schneider, G, Schnell, R, Agren, D. | | Deposit date: | 2008-10-10 | | Release date: | 2008-11-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 1.9 A structure of the signal receiver domain of the putative response regulator NarL from Mycobacterium tuberculosis.
Acta Crystallogr.,Sect.F, 64, 2008
|
|
6ZI3
 
 | | Crystal structure of OleP-6DEB bound to L-rhamnose | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 6-DEOXYERYTHRONOLIDE B, Cytochrome P-450, ... | | Authors: | Montemiglio, L.C, Savino, C, Vallone, B, Parisi, G, Freda, I. | | Deposit date: | 2020-06-24 | | Release date: | 2020-10-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Dissecting the Cytochrome P450 OleP Substrate Specificity: Evidence for a Preferential Substrate.
Biomolecules, 10, 2020
|
|
2IJ3
 
 | | Structure of the A264H mutant of cytochrome P450 BM3 | | Descriptor: | Cytochrome P450 BM3, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Toogood, H.S, Leys, D. | | Deposit date: | 2006-09-29 | | Release date: | 2006-11-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and spectroscopic characterization of P450 BM3 mutants with unprecedented P450 heme iron ligand sets. New heme ligation states influence conformational equilibria in P450 BM3.
J.Biol.Chem., 282, 2007
|
|
5ERD
 
 | | Crystal structure of human Desmoglein-2 ectodomain | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Brasch, J, Harrison, O.J, Shapiro, L. | | Deposit date: | 2015-11-13 | | Release date: | 2016-06-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis of adhesive binding by desmocollins and desmogleins.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
3EV1
 
 | | Crystal Structure of Ribonuclease A in 70% Hexanediol | | Descriptor: | HEXANE-1,6-DIOL, Ribonuclease pancreatic | | Authors: | Dechene, M, Wink, G, Smith, M, Swartz, P, Mattos, C. | | Deposit date: | 2008-10-12 | | Release date: | 2009-06-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Multiple solvent crystal structures of ribonuclease A: An assessment of the method
Proteins, 76, 2009
|
|
6ZL5
 
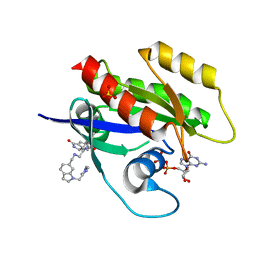 | | CRYSTAL STRUCTURE OF KRAS-G12D(C118S) IN COMPLEX WITH BI-2852 AND GDP | | Descriptor: | (3~{S})-3-[2-[[[1-[(1-methylimidazol-4-yl)methyl]indol-6-yl]methylamino]methyl]-1~{H}-indol-3-yl]-5-oxidanyl-2,3-dihydroisoindol-1-one, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Kessler, D, Fischer, G, Boettcher, J. | | Deposit date: | 2020-06-30 | | Release date: | 2020-08-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.645 Å) | | Cite: | Drugging all RAS isoforms with one pocket.
Future Med Chem, 12, 2020
|
|
6Z4C
 
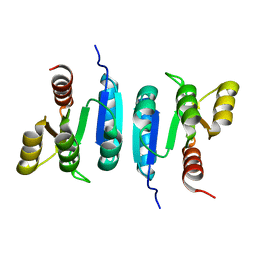 | | The structure of the N-terminal domain of RssB from E. coli | | Descriptor: | Regulator of RpoS | | Authors: | Zeth, K, Dimce, M, Terrence, D.M, Schuenemann, V, Dougan, D. | | Deposit date: | 2020-05-25 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Insight into the RssB-Mediated Recognition and Delivery of sigma s to the AAA+ Protease, ClpXP.
Biomolecules, 10, 2020
|
|
2IEY
 
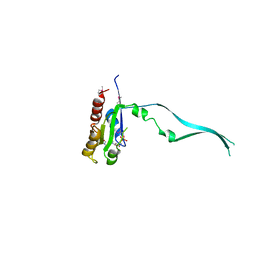 | | Crystal Structure of mouse Rab27b bound to GDP in hexagonal space group | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Ras-related protein Rab-27B | | Authors: | Chavas, L.M.G, Torii, S, Kamikubo, H, Kawasaki, M, Ihara, K, Kato, R, Kataoka, M, Izumi, T, Wakatsuki, S. | | Deposit date: | 2006-09-19 | | Release date: | 2007-05-01 | | Last modified: | 2012-04-11 | | Method: | X-RAY DIFFRACTION (3.18 Å) | | Cite: | Structure of the small GTPase Rab27b shows an unexpected swapped dimer
Acta Crystallogr.,Sect.D, 63, 2007
|
|
6Z4I
 
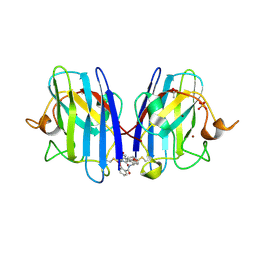 | |
2IKU
 
 | | Crystal Structure of Human Renin Complexed with Inhibitors | | Descriptor: | 6-ETHYL-5-[(2S)-1-(3-METHOXYPROPYL)-2-PHENYL-1,2,3,4-TETRAHYDROQUINOLIN-7-YL]PYRIMIDINE-2,4-DIAMINE, Renin | | Authors: | Mochalkin, I. | | Deposit date: | 2006-10-02 | | Release date: | 2006-12-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Binding thermodynamics of substituted diaminopyrimidine renin inhibitors.
Anal.Biochem., 360, 2007
|
|
6Z4O
 
 | |
6Z52
 
 | | Crystal structure of CLK3 in complex with macrocycle ODS2003136 | | Descriptor: | 1,2-ETHANEDIOL, 11,15-dimethyl-6-(4-methylpiperazin-1-yl)-8-oxa-2,11,15,19,21,23-hexazatetracyclo[15.6.1.13,7.020,24]pentacosa-1(23),3(25),4,6,17,20(24),21-heptaen-10-one, CHLORIDE ION, ... | | Authors: | Chaikuad, A, Benderitter, P, Hoflack, J, Denis, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-05-26 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Crystal structure of CLK3 in complex with macrocycle ODS2003136
To Be Published
|
|
5ER7
 
 | | Connexin-26 Bound to Calcium | | Descriptor: | CALCIUM ION, Gap junction beta-2 protein | | Authors: | Purdy, M.D, Bennett, B.C, Baker, K.A, Yeager, M.J. | | Deposit date: | 2015-11-13 | | Release date: | 2016-01-27 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (3.286 Å) | | Cite: | An electrostatic mechanism for Ca(2+)-mediated regulation of gap junction channels.
Nat Commun, 7, 2016
|
|
6Z67
 
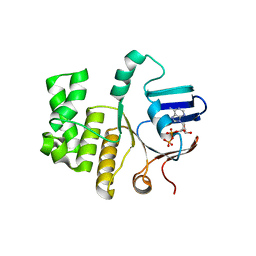 | | FtsE structure of Streptococcus pneumoniae in complex with AMPPNP at 2.4 A resolution | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cell division ATP-binding protein FtsE, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Alcorlo, M, Straume, D, Havarstein, L.S, Hermoso, j.A. | | Deposit date: | 2020-05-28 | | Release date: | 2020-09-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Characterization of the Essential Cell Division Protein FtsE and Its Interaction with FtsX in Streptococcus pneumoniae.
Mbio, 11, 2020
|
|
3EV2
 
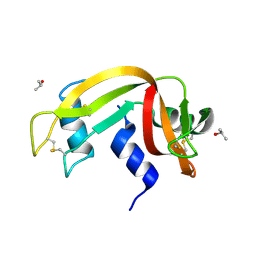 | | Crystal Structure of Ribonuclease A in 70% Isopropanol | | Descriptor: | ISOPROPYL ALCOHOL, Ribonuclease pancreatic | | Authors: | Dechene, M, Wink, G, Smith, M, Swartz, P, Mattos, C. | | Deposit date: | 2008-10-12 | | Release date: | 2009-06-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Multiple solvent crystal structures of ribonuclease A: An assessment of the method
Proteins, 76, 2009
|
|
6ZIR
 
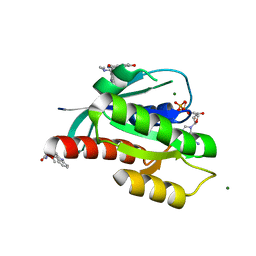 | | CRYSTAL STRUCTURE OF NRAS (C118S) IN COMPLEX WITH GDP AND COMPOUND 18 | | Descriptor: | (3~{S})-3-[2-[(dimethylamino)methyl]-1~{H}-indol-3-yl]-5-oxidanyl-2,3-dihydroisoindol-1-one, GTPase NRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Kessler, D, Fischer, G, Boettcher, J. | | Deposit date: | 2020-06-26 | | Release date: | 2020-08-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Drugging all RAS isoforms with one pocket.
Future Med Chem, 12, 2020
|
|
6ZIW
 
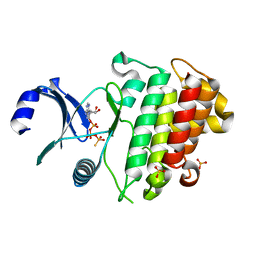 | | The IRAK3 Pseudokinase Domain Bound To ATPgammaS | | Descriptor: | Interleukin-1 receptor-associated kinase 3, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, SULFATE ION | | Authors: | Mathea, S, Chatterjee, D, Preuss, F, Kraemer, A, Knapp, S. | | Deposit date: | 2020-06-26 | | Release date: | 2020-07-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | The IRAK3 Pseudokinase Domain Bound To ATPgammaS
To Be Published
|
|
5ESF
 
 | | Saccharomyces cerevisiae CYP51 (Lanosterol 14-alpha demethylase) G73E mutant complexed with fluconazole | | Descriptor: | 2-(2,4-DIFLUOROPHENYL)-1,3-DI(1H-1,2,4-TRIAZOL-1-YL)PROPAN-2-OL, Lanosterol 14-alpha demethylase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sagatova, A, Keniya, M.V, Wilson, R.K, Sabherwal, M, Tyndall, J.D.A, Monk, B.C. | | Deposit date: | 2015-11-16 | | Release date: | 2016-11-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Impact of Homologous Resistance Mutations from Pathogenic Yeast on Saccharomyces cerevisiae Lanosterol 14 alpha-Demethylase.
Antimicrob.Agents Chemother., 62, 2018
|
|
3EV5
 
 | | Crystal Structure of Ribonuclease A in 1M Trimethylamine N-Oxide | | Descriptor: | Ribonuclease pancreatic, trimethylamine oxide | | Authors: | Dechene, M, Wink, G, Smith, M, Swartz, P, Mattos, C. | | Deposit date: | 2008-10-12 | | Release date: | 2009-06-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Multiple solvent crystal structures of ribonuclease A: An assessment of the method
Proteins, 76, 2009
|
|
3EGF
 
 | |
3ERK
 
 | | THE COMPLEX STRUCTURE OF THE MAP KINASE ERK2/SB220025 | | Descriptor: | 4-(4-FLUOROPHENYL)-1-(4-PIPERIDINYL)-5-(2-AMINO-4-PYRIMIDINYL)-IMIDAZOLE, EXTRACELLULAR REGULATED KINASE 2 | | Authors: | Wang, Z, Canagarajah, B, Boehm, J.C, Cobb, M.H, Young, P.R, Abdel-Meguid, S, Adams, J.L, Goldsmith, E.J. | | Deposit date: | 1998-07-09 | | Release date: | 1999-07-22 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of inhibitor selectivity in MAP kinases.
Structure, 6, 1998
|
|
3EST
 
 | | STRUCTURE OF NATIVE PORCINE PANCREATIC ELASTASE AT 1.65 ANGSTROMS RESOLUTION | | Descriptor: | CALCIUM ION, PORCINE PANCREATIC ELASTASE, SULFATE ION | | Authors: | Meyer, E.F, Cole, G, Radhakrishnan, R, Epp, O. | | Deposit date: | 1987-09-17 | | Release date: | 1988-01-16 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure of native porcine pancreatic elastase at 1.65 A resolutions.
Acta Crystallogr.,Sect.B, 44, 1988
|
|
