8TSC
 
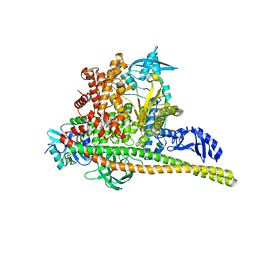 | | Human PI3K p85alpha/p110alpha H1047R bound to compound 3 | | Descriptor: | (1S)-7-[3-fluoro-5-(trifluoromethyl)benzamido]-N-methyl-1-(2-methylphenyl)-3-oxo-2,3-dihydro-1H-isoindole-5-carboxamide, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Holliday, M, Tang, Y, Bulku, A, Wilbur, J, Fraser, J. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.62 Å) | | Cite: | Discovery and Clinical Proof-of-Concept of RLY-2608, a First-in-Class Mutant-Selective Allosteric PI3K alpha Inhibitor That Decouples Antitumor Activity from Hyperinsulinemia.
Cancer Discov, 14, 2024
|
|
8UCD
 
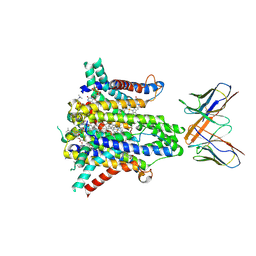 | | Cryo-EM structure of human STEAP1 in complex with AMG 509 Fab | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, AMG 509 anti-STEAP1 Fab, heavy chain, ... | | Authors: | Li, F, Bailis, J.M, Zhang, H. | | Deposit date: | 2023-09-26 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | AMG 509 (Xaluritamig), an Anti-STEAP1 XmAb 2+1 T-cell Redirecting Immune Therapy with Avidity-Dependent Activity against Prostate Cancer.
Cancer Discov, 14, 2024
|
|
8TUI
 
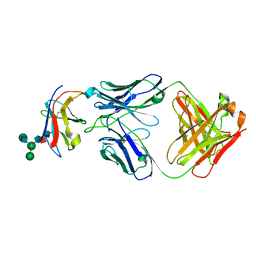 | | Crystal structure of Fab-Lirilumab bound to KIR2DL3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Killer cell immunoglobulin-like receptor 2DL3, Lirilumab Fab Heavy Chain, ... | | Authors: | Lorig-Roach, N, DuBois, R.M. | | Deposit date: | 2023-08-16 | | Release date: | 2023-12-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural basis for the activity and specificity of the immune checkpoint inhibitor lirilumab.
Sci Rep, 14, 2024
|
|
6ONM
 
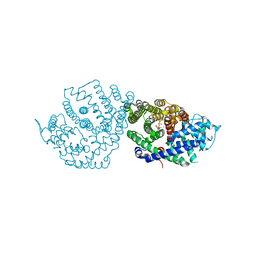 | | Crystal Structure of (+)-Limonene Synthase Complexed with 8,9-Difluorolinalyl Diphosphate | | Descriptor: | (+)-limonene synthase, (3R)-8-fluoro-7-(fluoromethyl)-3-methylocta-1,6-dien-3-yl trihydrogen diphosphate, MANGANESE (II) ION | | Authors: | Prem Kumar, R, Morehouse, B.R, Yu, Q, Oprian, D.D. | | Deposit date: | 2019-04-22 | | Release date: | 2019-09-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Direct Evidence of an Enzyme-Generated LPP Intermediate in (+)-Limonene Synthase Using a Fluorinated GPP Substrate Analog.
Acs Chem.Biol., 14, 2019
|
|
6OOI
 
 | |
7RM6
 
 | |
8U1U
 
 | | Structure of a class A GPCR/agonist complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, C-C motif chemokine 1,C-C chemokine receptor type 8,EGFP fusion protein, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Sun, D, Johnson, M, Masureel, M. | | Deposit date: | 2023-09-02 | | Release date: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of antibody inhibition and chemokine activation of the human CC chemokine receptor 8.
Nat Commun, 14, 2023
|
|
3T4N
 
 | | Structure of the regulatory fragment of Saccharomyces cerevisiae AMPK in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Carbon catabolite-derepressing protein kinase, Nuclear protein SNF4, ... | | Authors: | Mayer, F.V, Heath, R, Underwood, E, Sanders, M.J, Carmena, D, McCartney, R, Leiper, F.C, Xiao, B, Jing, C, Walker, P.A, Haire, L.F, Ogrodowicz, R, Martin, S.R, Schmdit, M.C, Gamblin, S.J, Carling, D. | | Deposit date: | 2011-07-26 | | Release date: | 2011-11-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | ADP Regulates SNF1, the Saccharomyces cerevisiae Homolog of AMP-Activated Protein Kinase.
Cell Metab, 14, 2011
|
|
8UGC
 
 | |
8U7X
 
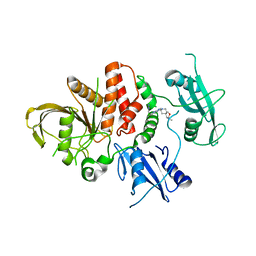 | |
6OPS
 
 | | HIV-1 Protease NL4-3 WT in complex with darunavir | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, Protease NL4-3 | | Authors: | Lockbaum, G.J, Henes, M, Kosovrasti, K, Leidner, F, Nachum, G.S, Nalivaika, E.A, Bolon, D.N.A, KurtYilmaz, N, Schiffer, C.A. | | Deposit date: | 2019-04-25 | | Release date: | 2019-09-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Picomolar to Micromolar: Elucidating the Role of Distal Mutations in HIV-1 Protease in Conferring Drug Resistance.
Acs Chem.Biol., 14, 2019
|
|
6OPW
 
 | | HIV-1 Protease NL4-3 I13V, G16E, V32I, L33F, K45I, M46I, A71V, V82F, I84V Mutant in complex with darunavir | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, Protease NL4-3, SULFATE ION | | Authors: | Lockbaum, G.J, Henes, M, Kosovrasti, K, Leidner, F, Nachum, G.S, Nalivaika, E.A, Bolon, D.N.A, KurtYilmaz, N, Schiffer, C.A. | | Deposit date: | 2019-04-25 | | Release date: | 2019-09-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Picomolar to Micromolar: Elucidating the Role of Distal Mutations in HIV-1 Protease in Conferring Drug Resistance.
Acs Chem.Biol., 14, 2019
|
|
3SQ9
 
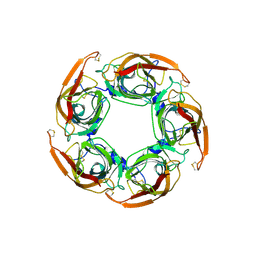 | | Crystal Structures of the Ligand Binding Domain of a Pentameric Alpha7 Nicotinic Receptor Chimera | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Neuronal acetylcholine receptor subunit alpha-7, ... | | Authors: | Li, S.-X, Huang, S, Bren, N, Noridomi, K, Dellisanti, C, Sine, S, Chen, L. | | Deposit date: | 2011-07-05 | | Release date: | 2011-09-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Ligand-binding domain of an alpha 7-nicotinic receptor chimera and its complex with agonist.
Nat.Neurosci., 14, 2011
|
|
4ODM
 
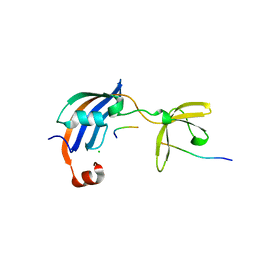 | | Structure of SlyD from Thermus thermophilus in complex with S2-W23A peptide | | Descriptor: | 30S ribosomal protein S2, ACETATE ION, CHLORIDE ION, ... | | Authors: | Quistgaard, E.M, Low, C, Nordlund, P. | | Deposit date: | 2014-01-10 | | Release date: | 2015-01-14 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Molecular insights into substrate recognition and catalytic mechanism of the chaperone and FKBP peptidyl-prolyl isomerase SlyD.
BMC Biol., 14, 2016
|
|
7R7Z
 
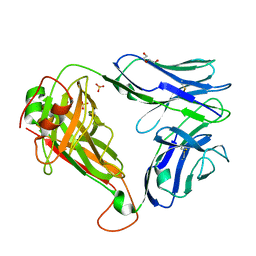 | | Crystal structure of QW9-HLA-B*5301 specific T Cell Receptor, C3 | | Descriptor: | Alpha chain of C3 TCR, Beta chain of C3 TCR, HEXAETHYLENE GLYCOL, ... | | Authors: | Li, X.L, Tan, K.M, Xu, S.T, Ng, R, Walker, B.D, Wang, J.H. | | Deposit date: | 2021-06-25 | | Release date: | 2022-06-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.286 Å) | | Cite: | Molecular basis of differential HLA class I-restricted T cell recognition of a highly networked HIV peptide.
Nat Commun, 14, 2023
|
|
7R7V
 
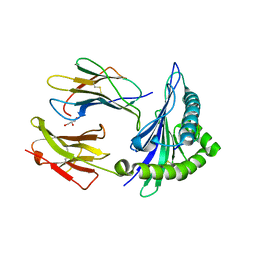 | | Crystal structure of HLA-B*5301 complex with an HIV-1 Gag-derived epitope QW9 | | Descriptor: | Beta-2-microglobulin, GLN-ALA-SER-GLN-GLU-VAL-LYS-ASN-TRP, GLYCEROL, ... | | Authors: | Li, X.L, Tan, K.M, Walker, B.D, Wang, J.H. | | Deposit date: | 2021-06-25 | | Release date: | 2022-06-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular basis of differential HLA class I-restricted T cell recognition of a highly networked HIV peptide.
Nat Commun, 14, 2023
|
|
7R7W
 
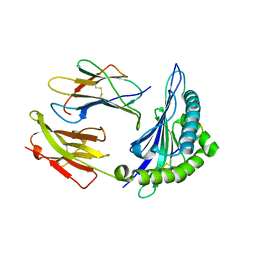 | | Crystal structure of HLA-B*5301 complex with an HIV-1 Gag-derived epitope QW9 S3T variant | | Descriptor: | Beta-2-microglobulin, GLN-ALA-THR-GLN-GLU-VAL-LYS-ASN-TRP, MHC class I antigen | | Authors: | Li, X.L, Tan, K.M, Walker, B.D, Wang, J.H. | | Deposit date: | 2021-06-25 | | Release date: | 2022-06-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Molecular basis of differential HLA class I-restricted T cell recognition of a highly networked HIV peptide.
Nat Commun, 14, 2023
|
|
7R7Y
 
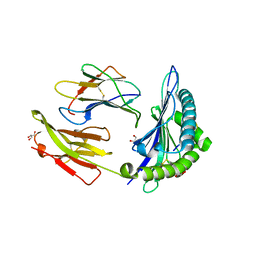 | | Crystal structure of HLA-B*5701 complex with an HIV-1 Gag-derived epitope QW9 S3T variant | | Descriptor: | Beta-2-microglobulin, GLN-ALA-THR-GLN-GLU-VAL-LYS-ASN-TRP, GLYCEROL, ... | | Authors: | Li, X.L, Ng, R, Tan, K.M, Walker, B.D, Wang, J.H. | | Deposit date: | 2021-06-25 | | Release date: | 2022-06-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Molecular basis of differential HLA class I-restricted T cell recognition of a highly networked HIV peptide.
Nat Commun, 14, 2023
|
|
7R7X
 
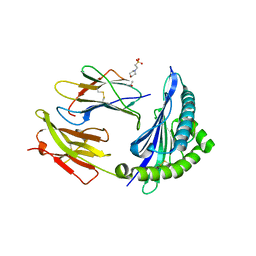 | | Crystal structure of HLA-B*5701 complex with an HIV-1 Gag-derived epitope QW9 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Beta-2-microglobulin, GLN-ALA-SER-GLN-GLU-VAL-LYS-ASN-TRP, ... | | Authors: | Li, X.L, Tan, K.M, Walker, B.D, Wang, J.H. | | Deposit date: | 2021-06-25 | | Release date: | 2022-06-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | Molecular basis of differential HLA class I-restricted T cell recognition of a highly networked HIV peptide.
Nat Commun, 14, 2023
|
|
7R80
 
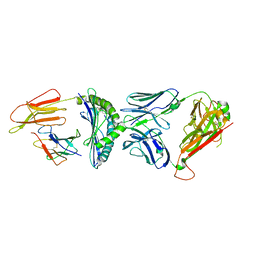 | | Crystal structure of C3 TCR complex with QW9-bound HLA-B*5301 | | Descriptor: | Alpha chain of C3 TCR, Beta Chain of C3 TCR, Beta-2-microglobulin, ... | | Authors: | Li, X.L, Tan, K.M, Walker, B.D, Wang, J.H. | | Deposit date: | 2021-06-25 | | Release date: | 2022-06-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Molecular basis of differential HLA class I-restricted T cell recognition of a highly networked HIV peptide.
Nat Commun, 14, 2023
|
|
3UPI
 
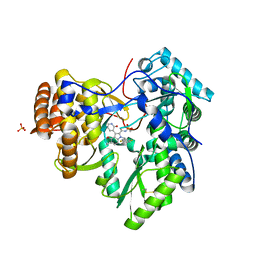 | | Synthesis of novel 4,5-dihydrofurano indoles and their evaluation as HCV NS5B polymerase inhibitors | | Descriptor: | (3S)-6-(2,5-difluorobenzyl)-3-methyl-N-(methylsulfonyl)-8-(2-oxo-1,2-dihydropyridin-3-yl)-3,6-dihydro-2H-furo[2,3-e]indole-7-carboxamide, PHOSPHATE ION, RNA-directed RNA polymerase | | Authors: | Velazquez, F, Venkataraman, S, Lesburg, C.A, Duca, J.S, Rosenblum, S.B, Kozlowski, J.A, Njoroge, F.G. | | Deposit date: | 2011-11-18 | | Release date: | 2012-01-25 | | Last modified: | 2012-02-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Synthesis of New 4,5-Dihydrofuranoindoles and Their Evaluation as HCV NS5B Polymerase Inhibitors.
Org.Lett., 14, 2012
|
|
8UAJ
 
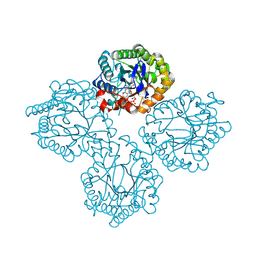 | | Succinate Bound Crystal Structure of Thermus scotoductus SA-01 Ene-reductase | | Descriptor: | FLAVIN MONONUCLEOTIDE, NADPH dehydrogenase, SUCCINIC ACID | | Authors: | Wilson, L.A, Guddat, L, Schenk, G, Scott, C. | | Deposit date: | 2023-09-21 | | Release date: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Structural Characterization of Enzymatic Interactions with Functional Nicotinamide Cofactor Biomimetics
Catalysts, 14, 2024
|
|
8UAS
 
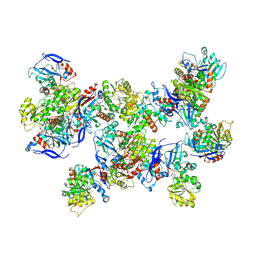 | | Rhodococcus ruber Alcohol Dehydrogenase NADH Biomimetic Complex - Compound 1a | | Descriptor: | 1-[3-[~{tert}-butyl(dimethyl)silyl]oxypropyl]pyridine-3-carboxamide, CITRIC ACID, ISOPROPYL ALCOHOL, ... | | Authors: | Wilson, L.A, Guddat, L.W, Schenk, G, Scott, C. | | Deposit date: | 2023-09-22 | | Release date: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Characterization of Enzymatic Interactions with Functional Nicotinamide Cofactor Biomimetics
Catalysts, 14, 2024
|
|
8UAR
 
 | | Rhodococcus ruber Alcohol Dehydrogenase NADH Biomimetic Complex - Compound 4b | | Descriptor: | 1-{[4-(hydroxymethyl)phenyl]methyl}-1,4-dihydropyridine-3-carboxamide, CITRIC ACID, ISOPROPYL ALCOHOL, ... | | Authors: | Wilson, L.A, Schenk, G, Guddat, L.W, Scott, C. | | Deposit date: | 2023-09-22 | | Release date: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Structural Characterization of Enzymatic Interactions with Functional Nicotinamide Cofactor Biomimetics
Catalysts, 14, 2024
|
|
8UAT
 
 | | Thermus scotoductus SA-01 Ene-reductase Compound 3b Complex | | Descriptor: | 1-[2-(4-hydroxyphenyl)ethyl]-1,4-dihydropyridine-3-carboxamide, CHLORIDE ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Wilson, L.A, Guddat, L.W, Schenk, G, Scott, C. | | Deposit date: | 2023-09-22 | | Release date: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Structural Characterization of Enzymatic Interactions with Functional Nicotinamide Cofactor Biomimetics
Catalysts, 14, 2024
|
|
