6II1
 
 | | Crystal Structure Analysis of CO form hemoglobin from Bos taurus | | Descriptor: | CARBON MONOXIDE, Hemoglobin subunit alpha, Hemoglobin subunit beta, ... | | Authors: | Kihira, K, Morita, Y, Yamada, T, Kureishi, M, Komatsu, T. | | Deposit date: | 2018-10-03 | | Release date: | 2018-12-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Quaternary Structure Analysis of a Hemoglobin Core in Hemoglobin-Albumin Cluster.
J Phys Chem B, 122, 2018
|
|
5X1S
 
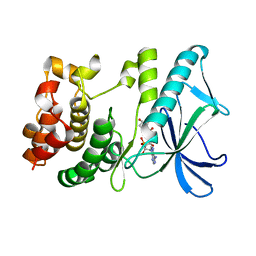 | | PpkA-294 with Amppcp | | Descriptor: | GLYCEROL, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, PpkA | | Authors: | Li, P.P, Ran, T.T, Xu, D.Q, Wang, W.W. | | Deposit date: | 2017-01-26 | | Release date: | 2018-01-31 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structures of the kinase domain of PpkA, a key regulatory component of T6SS, reveal a general inhibitory mechanism.
Biochem.J., 475, 2018
|
|
8E54
 
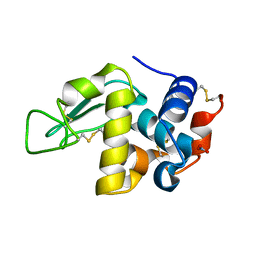 | | MicroED structure of triclinic lysozyme recorded on K3 | | Descriptor: | Lysozyme C, NITRATE ION | | Authors: | Clabbers, M.T.B, Martynowycz, M.W, Hattne, J, Nannenga, B.L, Gonen, T. | | Deposit date: | 2022-08-19 | | Release date: | 2022-09-21 | | Last modified: | 2022-10-19 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.2 Å) | | Cite: | Electron-counting MicroED data with the K2 and K3 direct electron detectors.
J.Struct.Biol., 214, 2022
|
|
6LJV
 
 | | Crystal structure of human FABP4 in complex with a novel inhibitor | | Descriptor: | 2-[[3-chloranyl-2-(2,3-dihydro-1-benzofuran-5-yl)phenyl]amino]benzoic acid, Fatty acid-binding protein, adipocyte | | Authors: | Su, H.X, Zhang, X.L, Li, M.J, Xu, Y.C. | | Deposit date: | 2019-12-17 | | Release date: | 2020-04-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.401 Å) | | Cite: | Exploration of Fragment Binding Poses Leading to Efficient Discovery of Highly Potent and Orally Effective Inhibitors of FABP4 for Anti-inflammation.
J.Med.Chem., 63, 2020
|
|
5WDZ
 
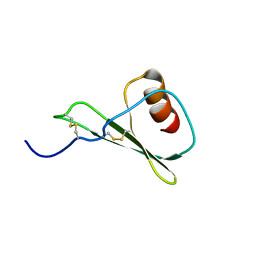 | |
6L14
 
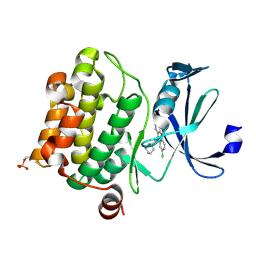 | | Crystal structure of Ser/Thr kinase Pim1 in complex with 10-DEBC derivatives | | Descriptor: | 2-chloranyl-10-[3-[(3~{S})-piperidin-3-yl]propyl]phenoxazine, Serine/threonine-protein kinase pim-1 | | Authors: | Zhang, W, Xie, Y, Cao, R, Huang, N, Zhou, Y. | | Deposit date: | 2019-09-27 | | Release date: | 2020-05-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-Based Optimization of 10-DEBC Derivatives as Potent and Selective Pim-1 Kinase Inhibitors.
J.Chem.Inf.Model., 60, 2020
|
|
8JAH
 
 | | Crystal structure of human CLEC12A C-type lectin domain | | Descriptor: | C-type lectin domain family 12 member A, POTASSIUM ION, SULFATE ION | | Authors: | Lei, Q, Tang, H, Dong, X. | | Deposit date: | 2023-05-06 | | Release date: | 2024-03-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Mechanistic insights into the C-type lectin receptor CLEC12A-mediated immune recognition of monosodium urate crystal.
J.Biol.Chem., 300, 2024
|
|
6XH3
 
 | |
6IN4
 
 | | Crystal structure of apo DAPK1 in the presence of 18-crown-6 | | Descriptor: | Death-associated protein kinase 1, SULFATE ION | | Authors: | Yokoyama, T, Kosaka, Y, Matsumoto, K, Kitakami, R, Nabeshima, Y, Mizuguchi, M. | | Deposit date: | 2018-10-24 | | Release date: | 2019-10-30 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crown Ethers as Transthyretin Amyloidogenesis Inhibitor
To Be Published
|
|
5X2U
 
 | |
6IPY
 
 | |
8IYX
 
 | | Cryo-EM structure of the GPR34 receptor in complex with the antagonist YL-365 | | Descriptor: | 1-[4-(3-chlorophenyl)phenyl]carbonyl-4-[2-(4-phenylmethoxyphenyl)ethanoylamino]piperidine-4-carboxylic acid, Probable G-protein coupled receptor 34,YL-365 | | Authors: | Jia, G.W, Wang, X, Zhang, C.B, Dong, H.H, Su, Z.M. | | Deposit date: | 2023-04-06 | | Release date: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | Cryo-EM structures of human GPR34 enable the identification of selective antagonists.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
5X40
 
 | | Structure of a CbiO dimer bound with AMPPCP | | Descriptor: | Cobalt ABC transporter ATP-binding protein, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER | | Authors: | Bao, Z, Qi, X, Wang, J, Zhang, P. | | Deposit date: | 2017-02-09 | | Release date: | 2017-04-05 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure and mechanism of a group-I cobalt energy coupling factor transporter
Cell Res., 27, 2017
|
|
8DS5
 
 | | X-ray structure of the MK5890 Fab - CD27 antibody-antigen complex | | Descriptor: | CADMIUM ION, CD27 antigen, MK-5890 Fab heavy chain, ... | | Authors: | Fischmann, T.O. | | Deposit date: | 2022-07-21 | | Release date: | 2022-09-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.926 Å) | | Cite: | Preclinical characterization and clinical translation of pharmacodynamic markers for MK-5890: a human CD27 activating antibody for cancer immunotherapy.
J Immunother Cancer, 10, 2022
|
|
8V52
 
 | |
8UDC
 
 | | Crystal structure of TcPINK1 in complex with CYC116 | | Descriptor: | (4P)-4-(2-amino-4-methyl-1,3-thiazol-5-yl)-N-[4-(morpholin-4-yl)phenyl]pyrimidin-2-amine, DI(HYDROXYETHYL)ETHER, SULFATE ION, ... | | Authors: | Veyron, S, Rasool, S, Trempe, J.F. | | Deposit date: | 2023-09-28 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural Characterization of a small-molecule inhibitor of PINK1, a precursor tool compound for the study of Parkinson's disease
To Be Published
|
|
5X94
 
 | | Crystal structure of SHP2_SH2-CagA EPIYA_D peptide complex | | Descriptor: | Cag pathogenicity island protein, Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Senda, M, Senda, T. | | Deposit date: | 2017-03-05 | | Release date: | 2017-09-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Differential Mechanisms for SHP2 Binding and Activation Are Exploited by Geographically Distinct Helicobacter pylori CagA Oncoproteins.
Cell Rep, 20, 2017
|
|
6IS2
 
 | |
6LAP
 
 | | Cryo-EM structure of echovirus 11 A-particle at pH 7.4 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3 | | Authors: | Liu, S, Gao, F.G. | | Deposit date: | 2019-11-13 | | Release date: | 2020-10-07 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.49 Å) | | Cite: | Molecular and structural basis of Echovirus 11 infection by using the dual-receptor system of CD55 and FcRn.
Chin.Sci.Bull., 2020
|
|
6LCX
 
 | | Crosslinked alpha(Ni)-beta(Ni) human hemoglobin A in the T quaternary structure at 95 K: Light | | Descriptor: | BUT-2-ENEDIAL, Hemoglobin subunit alpha, Hemoglobin subunit beta, ... | | Authors: | Shibayama, N, Park, S.Y, Ohki, M, Sato-Tomita, A. | | Deposit date: | 2019-11-20 | | Release date: | 2020-02-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Direct observation of ligand migration within human hemoglobin at work.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6I19
 
 | | Crystal structure of Chlamydomonas reinhardtii thioredoxin h1 | | Descriptor: | Thioredoxin H-type | | Authors: | Lemaire, S.D, Tedesco, D, Crozet, P, Michelet, L, Fermani, S, Zaffagnini, M, Henri, J. | | Deposit date: | 2018-10-27 | | Release date: | 2018-12-05 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.378 Å) | | Cite: | Crystal Structure of Chloroplastic Thioredoxin f2 fromChlamydomonas reinhardtiiReveals Distinct Surface Properties.
Antioxidants (Basel), 7, 2018
|
|
6I3I
 
 | |
5XED
 
 | | Heterodimer constructed from M61A PA cyt c551-HT cyt c552 and HT cyt c552-PA cyt c551 chimeric proteins | | Descriptor: | Cytochrome c-551,Cytochrome c-552, Cytochrome c-552,Cytochrome c-551, HEME C | | Authors: | Zhang, M, Nakanishi, T, Yamanaka, M, Nagao, S, Yanagisawa, S, Shomura, Y, Shibata, N, Ogura, T, Higuchi, Y, Hirota, S. | | Deposit date: | 2017-04-04 | | Release date: | 2017-08-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Rational Design of Domain-Swapping-Based c-Type Cytochrome Heterodimers by Using Chimeric Proteins.
Chembiochem, 18, 2017
|
|
5X2R
 
 | |
8V2Z
 
 | | Cryo-EM Structure of Smooth Muscle Gamma Actin (ACTG2) Mutant R257C | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, gamma-enteric smooth muscle, ... | | Authors: | Palmer, N.J, Carman, P.J, Ceron, R.H, Dominguez, R. | | Deposit date: | 2023-11-25 | | Release date: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.72 Å) | | Cite: | Smooth Muscle Gamma Actin (ACTG2) Filament Mutant R257C
To Be Published
|
|
