3E8K
 
 | |
3E9F
 
 | | Crystal structure short-form (residue1-113) of Eaf3 chromo domain | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Chromatin modification-related protein EAF3 | | Authors: | Sun, B, Hong, J, Zhang, P, Lin, D, Ding, J. | | Deposit date: | 2008-08-22 | | Release date: | 2008-11-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular Basis of the Interaction of Saccharomyces cerevisiae Eaf3 Chromo Domain with Methylated H3K36
J.Biol.Chem., 283, 2008
|
|
3V0L
 
 | | Crystal structure of the Fucosylgalactoside alpha N-acetylgalactosaminyltransferase (GTA, cisAB mutant L266G, G268A) in complex with a novel UDP-Gal derived inhibitor (2GW) | | Descriptor: | 5-phenyl-uridine-5'-alpha-d-galactosyl-diphosphate, Histo-blood group ABO system transferase, MANGANESE (II) ION, ... | | Authors: | Palcic, M.M, Jorgensen, R. | | Deposit date: | 2011-12-08 | | Release date: | 2013-01-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Base-modified Donor Analogues Reveal Novel Dynamic Features of a Glycosyltransferase.
J.Biol.Chem., 288, 2013
|
|
3MV6
 
 | |
3E9Z
 
 | | Crystal structure of purine nucleoside phosphorylase from Schistosoma mansoni in complex with 6-chloroguanine | | Descriptor: | 6-chloroguanine, ACETATE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Pereira, H.M, Rezende, M.M, Oliva, G, Garratt, R.C. | | Deposit date: | 2008-08-24 | | Release date: | 2009-09-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Crystal structure of Schistosoma mansoni purine nucleoside phosphorylase (SmPNP) in complex with adenine, 8-aminoguanine, 8-azaguanine and 6-chloroguanine.
To be Published
|
|
3EAI
 
 | | Structure of inhibited murine iNOS oxygenase domain | | Descriptor: | 4-({4-[(4-methoxypyridin-2-yl)amino]piperidin-1-yl}carbonyl)benzonitrile, 5,6,7,8-TETRAHYDROBIOPTERIN, Nitric oxide synthase, ... | | Authors: | Garcin, E.D, Arvai, A.S, Rosenfeld, R.J, Kroeger, M.D, Crane, B.R, Andersson, G, Andrews, G, Hamley, P.J, Mallinder, P.R, Nicholls, D.J, St-Gallay, S.A, Tinker, A.C, Gensmantel, N.P, Mete, A, Cheshire, D.R, Connolly, S, Stuehr, D.J, Aberg, A, Wallace, A.V, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2008-08-25 | | Release date: | 2008-10-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Anchored plasticity opens doors for selective inhibitor design in nitric oxide synthase.
Nat.Chem.Biol., 4, 2008
|
|
3V1N
 
 | | Crystal Structure of the H265Q mutant of a C-C hydrolase, BphD from Burkholderia xenovorans LB400, after exposure to its substrate HOPDA | | Descriptor: | (3E)-2,6-DIOXO-6-PHENYLHEX-3-ENOATE, 2-hydroxy-6-oxo-6-phenylhexa-2,4-dienoate hydrolase, BENZOIC ACID, ... | | Authors: | Ghosh, S, Bolin, J.T. | | Deposit date: | 2011-12-09 | | Release date: | 2012-03-21 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Identification of an Acyl-Enzyme Intermediate in a meta-Cleavage Product Hydrolase Reveals the Versatility of the Catalytic Triad.
J.Am.Chem.Soc., 134, 2012
|
|
3V1T
 
 | |
4V5W
 
 | | Grapevine Fanleaf virus | | Descriptor: | COAT PROTEIN | | Authors: | Schellenberger, P, Demangeat, G, Ritzenthaler, C, Lorber, B, Sauter, C. | | Deposit date: | 2011-05-10 | | Release date: | 2014-07-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Exploiting Protein Engineering and Crystal Polymorphism for Successful X-Ray Structure Determination
Cryst.Growth Des., 11, 2011
|
|
3E2I
 
 | | Crystal structure of Thymidine Kinase from S. aureus | | Descriptor: | GLYCEROL, Thymidine kinase, ZINC ION | | Authors: | Lam, R, Johns, K, Battaile, K.P, Romanov, V, Lam, K, Pai, E.F, Chirgadze, N.Y. | | Deposit date: | 2008-08-05 | | Release date: | 2009-08-11 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal structure of Thymidine Kinase from S. aureus
To be Published
|
|
3V78
 
 | | Crystal Structure of Transcriptional Regulator | | Descriptor: | ETHIDIUM, PROBABLE TRANSCRIPTIONAL REGULATORY PROTEIN (PROBABLY DEOR-FAMILY) | | Authors: | Do, S.V, Bolla, J.R, Chen, X, Yu, E.W. | | Deposit date: | 2011-12-20 | | Release date: | 2012-12-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.299 Å) | | Cite: | Structural and functional analysis of the transcriptional regulator Rv3066 of Mycobacterium tuberculosis.
Nucleic Acids Res., 40, 2012
|
|
6NCF
 
 | | The structure of Stable-5-Lipoxygenase bound to AKBA | | Descriptor: | (3alpha,8alpha,17alpha,18alpha)-3-(acetyloxy)-11-oxours-12-en-23-oic acid, Arachidonate 5-lipoxygenase, FE (II) ION | | Authors: | Newcomer, M.E, Gilbert, N.C, Neau, D.B. | | Deposit date: | 2018-12-11 | | Release date: | 2020-05-13 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.871 Å) | | Cite: | Structural and mechanistic insights into 5-lipoxygenase inhibition by natural products.
Nat.Chem.Biol., 16, 2020
|
|
3MYT
 
 | | Crystal structure of Ketosteroid Isomerase D38HD99N from Pseudomonas testosteroni (tKSI) | | Descriptor: | EQUILENIN, GLYCEROL, SULFATE ION, ... | | Authors: | Gonzalez, A, Tsai, Y, Schwans, J, Sunden, F, Herschlag, D. | | Deposit date: | 2010-05-11 | | Release date: | 2011-11-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.961 Å) | | Cite: | Crystal Structure of Ketosteroid Isomerase D38HD99N from Pseudomonas Testosteroni (tKSI)
To be Published
|
|
3EBI
 
 | | Structure of the M1 Alanylaminopeptidase from malaria complexed with the phosphinate dipeptide analog | | Descriptor: | (2S)-3-[(R)-[(1S)-1-amino-3-phenylpropyl](hydroxy)phosphoryl]-2-benzylpropanoic acid, GLYCEROL, M1 family aminopeptidase, ... | | Authors: | McGowan, S, Porter, C.J, Buckle, A.M, Whisstock, J.C. | | Deposit date: | 2008-08-27 | | Release date: | 2009-01-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the inhibition of the essential Plasmodium falciparum M1 neutral aminopeptidase
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3MZC
 
 | | Human carbonic ahydrase II in complex with a benzenesulfonamide inhibitor | | Descriptor: | 4-[(cyclopentylcarbamoyl)amino]benzenesulfonamide, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Avvaru, B.S, Wagner, J, Robbins, A.H, Mckenna, R. | | Deposit date: | 2010-05-12 | | Release date: | 2011-03-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.498 Å) | | Cite: | Selective hydrophobic pocket binding observed within the carbonic anhydrase II active site accommodate different 4-substituted-ureido-benzenesulfonamides and correlate to inhibitor potency.
Chem.Commun.(Camb.), 46, 2010
|
|
6DHW
 
 | |
3V3Q
 
 | | Crystal Structure of Human Nur77 Ligand-binding Domain in Complex with Ethyl 2-[2,3,4 trimethoxy-6(1-octanoyl)phenyl]acetate | | Descriptor: | GLYCEROL, Nuclear receptor subfamily 4 group A member 1, SODIUM ION, ... | | Authors: | Zhang, Q, Shi, C, Yang, K, Chen, Y, Zhan, Y, Wu, Q, Lin, T. | | Deposit date: | 2011-12-14 | | Release date: | 2012-09-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | The orphan nuclear receptor Nur77 regulates LKB1 localization and activates AMPK
Nat.Chem.Biol., 8, 2012
|
|
3ECD
 
 | |
3N0W
 
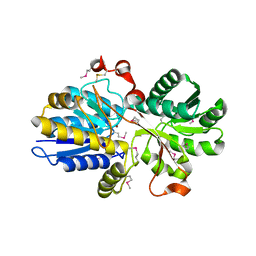 | |
3EDE
 
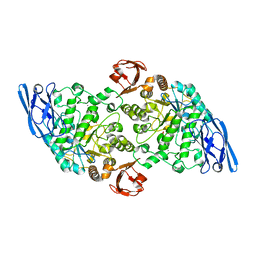 | |
4V6Y
 
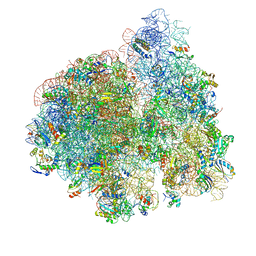 | | E. coli 70S-fMetVal-tRNAVal-tRNAfMet complex in classic pre-translocation state (pre1a) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Blau, C, Bock, L.V, Schroder, G.F, Davydov, I, Fischer, N, Stark, H, Rodnina, M.V, Vaiana, A.C, Grubmuller, H. | | Deposit date: | 2013-10-14 | | Release date: | 2014-07-09 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | Energy barriers and driving forces in tRNA translocation through the ribosome.
Nat.Struct.Mol.Biol., 20, 2013
|
|
3V4S
 
 | | Crystal Structure of ADP-ATP complex of purK: N5-carboxyaminoimidazole ribonucleotide synthetase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, CARBONATE ION, ... | | Authors: | Fung, L.W, Tuntland, M.L, Santarsiero, B.D, Johnson, M.E. | | Deposit date: | 2011-12-15 | | Release date: | 2013-02-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Elucidation of the bicarbonate binding site and insights into the carboxylation mechanism of (N(5))-carboxyaminoimidazole ribonucleotide synthase (PurK) from Bacillus anthracis.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4V7C
 
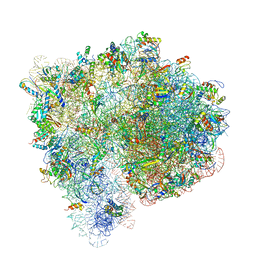 | | Structure of the Ribosome with Elongation Factor G Trapped in the Pre-Translocation State (pre-translocation 70S*tRNA structure) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Brilot, A.F, Korostelev, A.A, Ermolenko, D.N, Grigorieff, N. | | Deposit date: | 2013-11-20 | | Release date: | 2014-07-09 | | Last modified: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (7.6 Å) | | Cite: | Structure of the ribosome with elongation factor G trapped in the pretranslocation state.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3N37
 
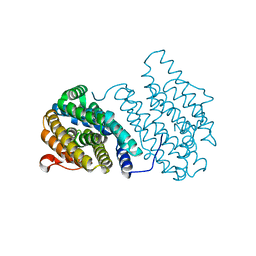 | | Ribonucleotide Reductase Dimanganese(II)-NrdF from Escherichia coli | | Descriptor: | GLYCEROL, MANGANESE (II) ION, Ribonucleoside-diphosphate reductase 2 subunit beta | | Authors: | Boal, A.K, Cotruvo Jr, J.A, Stubbe, J, Rosenzweig, A.C. | | Deposit date: | 2010-05-19 | | Release date: | 2010-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural basis for activation of class Ib ribonucleotide reductase.
Science, 329, 2010
|
|
3E6R
 
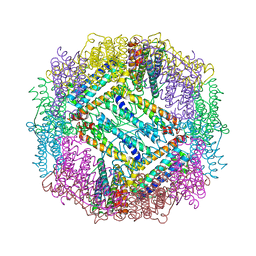 | |
