6WOO
 
 | | CryoEM structure of yeast 80S ribosome with Met-tRNAiMet, eIF5B, and GDP | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 5.8S ribosomal rRNA, ... | | Authors: | Wang, J, Wang, J, Puglisi, J, Fernandez, I.S. | | Deposit date: | 2020-04-25 | | Release date: | 2020-09-23 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | An active role of the eukaryotic large ribosomal subunit in translation initiation fidelity.
To Be Published
|
|
8I9Y
 
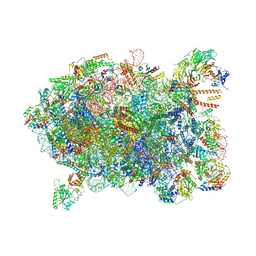 | | Cryo-EM structure of a Chaetomium thermophilum pre-60S ribosomal subunit - Ytm1-2 | | Descriptor: | 60S ribosomal protein L12-like protein, 60S ribosomal protein L13, 60S ribosomal protein L14-like protein, ... | | Authors: | Lau, B, Huang, Z, Beckmann, R, Hurt, E, Cheng, J. | | Deposit date: | 2023-02-07 | | Release date: | 2023-05-17 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Mechanism of 5S RNP recruitment and helicase-surveilled rRNA maturation during pre-60S biogenesis.
Embo Rep., 24, 2023
|
|
1CY6
 
 | |
1CLO
 
 | |
8OHN
 
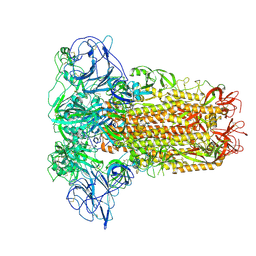 | | Human Coronavirus HKU1 spike glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Pronker, M.F, Hurdiss, D.L. | | Deposit date: | 2023-03-21 | | Release date: | 2023-08-02 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Sialoglycan binding triggers spike opening in a human coronavirus.
Nature, 624, 2023
|
|
1CLY
 
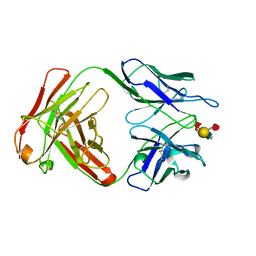 | |
8OPM
 
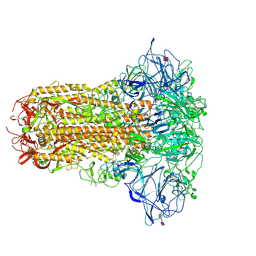 | | Human Coronavirus HKU1 spike glycoprotein in complex with an alpha2,8-linked 9-O-acetylated disialoside (closed state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 9-O-acetyl-5-acetamido-3,5-dideoxy-D-glycero-alpha-D-galacto-non-2-ulopyranosonic acid-(2-8)-N-acetyl-alpha-neuraminic acid, ... | | Authors: | Pronker, M.F, Creutznacher, R, Hurdiss, D.L. | | Deposit date: | 2023-04-07 | | Release date: | 2023-08-02 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Sialoglycan binding triggers spike opening in a human coronavirus.
Nature, 624, 2023
|
|
8IOS
 
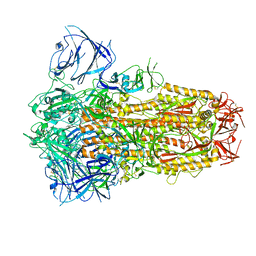 | | Structure of the SARS-CoV-2 XBB.1 spike glycoprotein (closed-1 state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Anraku, Y, Kita, S, Yajima, H, Sasaki, J, Sasaki-Tabata, K, Maenaka, K, Hashiguchi, T. | | Deposit date: | 2023-03-13 | | Release date: | 2023-05-24 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Virological characteristics of the SARS-CoV-2 XBB variant derived from recombination of two Omicron subvariants.
Nat Commun, 14, 2023
|
|
6WXG
 
 | | Cryo-EM reconstruction of VP5*/VP8* assembly from rhesus rotavirus particles - Reversed conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Intermediate capsid protein VP6, ... | | Authors: | Herrmann, T, Harrison, S.C, Jenni, S. | | Deposit date: | 2020-05-10 | | Release date: | 2021-01-20 | | Last modified: | 2021-03-10 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Functional refolding of the penetration protein on a non-enveloped virus.
Nature, 590, 2021
|
|
1CMX
 
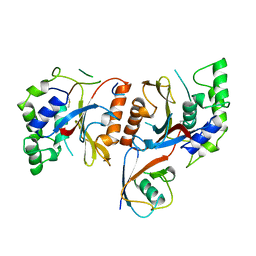 | |
8IOU
 
 | | Structure of SARS-CoV-2 XBB.1 spike glycoprotein in complex with ACE2 (1-up state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Anraku, Y, Kita, S, Yajima, H, Sasaki, J, Sasaki-Tabata, K, Maenaka, K, Hashiguchi, T. | | Deposit date: | 2023-03-13 | | Release date: | 2023-05-24 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Virological characteristics of the SARS-CoV-2 XBB variant derived from recombination of two Omicron subvariants.
Nat Commun, 14, 2023
|
|
1CQN
 
 | |
6X5K
 
 | | Crystal structure of CODH/ACS with carbon monoxide bound to the A-cluster | | Descriptor: | 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, CARBON MONOXIDE, Carbon monoxide dehydrogenase/acetyl-CoA synthase subunit alpha, ... | | Authors: | Cohen, S.E, Wittenborn, E.C, Hendrickson, R, Drennan, C.L. | | Deposit date: | 2020-05-26 | | Release date: | 2020-10-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Crystallographic Characterization of the Carbonylated A-Cluster in Carbon Monoxide Dehydrogenase/Acetyl-CoA Synthase
Acs Catalysis, 10, 2020
|
|
8IOT
 
 | | Structure of the SARS-CoV-2 XBB.1 spike glycoprotein (closed-2 state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Anraku, Y, Kita, S, Yajima, H, Sasaki, J, Sasaki-Tabata, K, Maenaka, K, Hashiguchi, T. | | Deposit date: | 2023-03-13 | | Release date: | 2023-05-24 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (2.51 Å) | | Cite: | Virological characteristics of the SARS-CoV-2 XBB variant derived from recombination of two Omicron subvariants.
Nat Commun, 14, 2023
|
|
1CR7
 
 | | PEANUT LECTIN-LACTOSE COMPLEX MONOCLINIC FORM | | Descriptor: | CALCIUM ION, LECTIN, MANGANESE (II) ION, ... | | Authors: | Ravishankar, R, Suguna, K, Surolia, A, Vijayan, M. | | Deposit date: | 1999-08-14 | | Release date: | 2001-04-21 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of the peanut lectin-lactose complex at acidic pH: retention of unusual quaternary structure, empty and carbohydrate bound combining sites, molecular mimicry and crystal packing directed by interactions at the combining site.
Proteins, 43, 2001
|
|
1D6U
 
 | | CRYSTAL STRUCTURE OF E. COLI AMINE OXIDASE ANAEROBICALLY REDUCED WITH BETA-PHENYLETHYLAMINE | | Descriptor: | 2-PHENYLETHYLAMINE, CALCIUM ION, COPPER (II) ION, ... | | Authors: | Wilmot, C.M, Hajdu, J, McPherson, M.J, Knowles, P.F, Phillips, S.E.V. | | Deposit date: | 1999-10-15 | | Release date: | 2000-02-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Visualization of dioxygen bound to copper during enzyme catalysis.
Science, 286, 1999
|
|
1CGT
 
 | |
1CUZ
 
 | | CUTINASE, L81G, L182G MUTANT | | Descriptor: | CUTINASE | | Authors: | Nicolas, A, Cambillau, C. | | Deposit date: | 1995-11-16 | | Release date: | 1996-07-11 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Dynamics of Fusarium solani cutinase investigated through structural comparison among different crystal forms of its variants.
Proteins, 26, 1996
|
|
6WD9
 
 | | Cryo-EM of elongating ribosome with EF-Tu*GTP elucidates tRNA proofreading (Cognate Structure III-B) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Loveland, A.B, Demo, G, Korostelev, A.A. | | Deposit date: | 2020-03-31 | | Release date: | 2020-07-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM of elongating ribosome with EF-Tu•GTP elucidates tRNA proofreading.
Nature, 584, 2020
|
|
1CDZ
 
 | | BRCT DOMAIN FROM DNA-REPAIR PROTEIN XRCC1 | | Descriptor: | PROTEIN (DNA-REPAIR PROTEIN XRCC1) | | Authors: | Zhang, X, Morera, S, Bates, P, Whitehead, P, Coffer, A, Hainbucher, K, Nash, R, Sternberg, M, Lindahl, T, Freemont, P. | | Deposit date: | 1999-03-04 | | Release date: | 2000-02-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of an XRCC1 BRCT domain: a new protein-protein interaction module.
EMBO J., 17, 1998
|
|
8OS6
 
 | | Structure of a GFRA1/GDNF LICAM complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Houghton, F.M, Adams, S.E, Briggs, D.C, McDonald, N.Q. | | Deposit date: | 2023-04-18 | | Release date: | 2023-11-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Architecture and regulation of a GDNF-GFR alpha 1 synaptic adhesion assembly.
Nat Commun, 14, 2023
|
|
1CVR
 
 | |
1CW9
 
 | | DNA DECAMER WITH AN ENGINEERED CROSSLINK IN THE MINOR GROOVE | | Descriptor: | 5'-D(*CP*CP*AP*GP*(G47)P*CP*CP*TP*GP*G)-3', CALCIUM ION | | Authors: | van Aalten, D.M.F, Erlanson, D.A, Verdine, G.L, Joshua-Tor, L. | | Deposit date: | 1999-08-26 | | Release date: | 1999-10-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A structural snapshot of base-pair opening in DNA.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1CH0
 
 | | RNASE T1 VARIANT WITH ALTERED GUANINE BINDING SEGMENT | | Descriptor: | CALCIUM ION, CHLORIDE ION, GUANOSINE-2'-MONOPHOSPHATE, ... | | Authors: | Hoeschler, K, Hoier, H, Orth, P, Hubner, B, Saenger, W, Hahn, U. | | Deposit date: | 1999-03-30 | | Release date: | 1999-12-22 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural analysis of an RNase T1 variant with an altered guanine binding segment.
J.Mol.Biol., 294, 1999
|
|
8OLL
 
 | | Staphylococcus aureus ClpP in complex with the natural product beta-lactone inhibitor Cystargolide A at 2.7 A resolution | | Descriptor: | ATP-dependent Clp protease proteolytic subunit, Cystargolide A (bound) | | Authors: | Illigmann, A, Vielberg, M.-T, Lakemeyer, M, Wolf, F, Staudt, N, Dema, T, Stange, P, Malik, I, Grond, S, Sieber, S.A, Groll, M, Kaysser, L, Broetz-Oesterhelt, H. | | Deposit date: | 2023-03-30 | | Release date: | 2023-12-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of Staphylococcus aureus ClpP Bound to the Covalent Active-Site Inhibitor Cystargolide A.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
