1XL6
 
 | | Intermediate gating structure 2 of the inwardly rectifying K+ channel KirBac3.1 | | Descriptor: | Inward rectifier potassium channel, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Gulbis, J.M, Kuo, A, Smith, B, Doyle, D.A, Edwards, A, Arrowsmith, C, Sundstrom, M. | | Deposit date: | 2004-09-30 | | Release date: | 2004-12-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Two intermediate gating state crystal structures of the KirBac3.1 K+ channel
To be Published
|
|
1XL7
 
 | | Crystal Structure of Mouse Carnitine Octanoyltransferase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Peroxisomal carnitine O-octanoyltransferase | | Authors: | Jogl, G, Hsiao, Y.S, Tong, L. | | Deposit date: | 2004-09-30 | | Release date: | 2004-10-19 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of mouse carnitine octanoyltransferase and molecular determinants of substrate selectivity.
J.Biol.Chem., 280, 2005
|
|
1XL8
 
 | | Crystal structure of mouse carnitine octanoyltransferase in complex with octanoylcarnitine | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CARNITINE, OCTANOYLCARNITINE, ... | | Authors: | Jogl, G, Hsiao, Y.S, Tong, L. | | Deposit date: | 2004-09-30 | | Release date: | 2004-10-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of mouse carnitine octanoyltransferase and molecular determinants of substrate selectivity.
J.Biol.Chem., 280, 2005
|
|
1XL9
 
 | | Crystal Structure of Dihydrodipicolinate Synthase DapA-2 (BA3935) from Bacillus Anthracis. | | Descriptor: | dihydrodipicolinate synthase | | Authors: | Blagova, E, Levdikov, V, Milioti, N, Fogg, M.J, Kalliomaa, A.K, Brannigan, J.A, Wilson, K.S, Wilkinson, A.J. | | Deposit date: | 2004-09-30 | | Release date: | 2005-10-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Crystal structure of dihydrodipicolinate synthase (BA3935) from Bacillus anthracis at 1.94 A resolution.
Proteins, 62, 2006
|
|
1XLA
 
 | |
1XLB
 
 | | MECHANISM FOR ALDOSE-KETOSE INTERCONVERSION BY D-XYLOSE ISOMERASE INVOLVING RING OPENING FOLLOWED BY A 1,2-HYDRIDE SHIFT | | Descriptor: | D-XYLOSE ISOMERASE, MAGNESIUM ION | | Authors: | Collyer, C.A, Henrick, K, Blow, D.M. | | Deposit date: | 1991-10-09 | | Release date: | 1993-07-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanism for aldose-ketose interconversion by D-xylose isomerase involving ring opening followed by a 1,2-hydride shift.
J.Mol.Biol., 212, 1990
|
|
1XLC
 
 | | MECHANISM FOR ALDOSE-KETOSE INTERCONVERSION BY D-XYLOSE ISOMERASE INVOLVING RING OPENING FOLLOWED BY A 1,2-HYDRIDE SHIFT | | Descriptor: | D-XYLOSE ISOMERASE, MAGNESIUM ION, Xylitol | | Authors: | Collyer, C.A, Henrick, K, Blow, D.M. | | Deposit date: | 1991-10-09 | | Release date: | 1993-07-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanism for aldose-ketose interconversion by D-xylose isomerase involving ring opening followed by a 1,2-hydride shift.
J.Mol.Biol., 212, 1990
|
|
1XLD
 
 | | MECHANISM FOR ALDOSE-KETOSE INTERCONVERSION BY D-XYLOSE ISOMERASE INVOLVING RING OPENING FOLLOWED BY A 1,2-HYDRIDE SHIFT | | Descriptor: | D-XYLOSE ISOMERASE, MANGANESE (II) ION, Xylitol | | Authors: | Collyer, C.A, Henrick, K, Blow, D.M. | | Deposit date: | 1991-10-09 | | Release date: | 1993-07-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanism for aldose-ketose interconversion by D-xylose isomerase involving ring opening followed by a 1,2-hydride shift.
J.Mol.Biol., 212, 1990
|
|
1XLE
 
 | | MECHANISM FOR ALDOSE-KETOSE INTERCONVERSION BY D-XYLOSE ISOMERASE INVOLVING RING OPENING FOLLOWED BY A 1,2-HYDRIDE SHIFT | | Descriptor: | D-XYLOSE ISOMERASE, MANGANESE (II) ION | | Authors: | Collyer, C.A, Henrick, K, Blow, D.M. | | Deposit date: | 1991-10-09 | | Release date: | 1993-07-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanism for aldose-ketose interconversion by D-xylose isomerase involving ring opening followed by a 1,2-hydride shift.
J.Mol.Biol., 212, 1990
|
|
1XLF
 
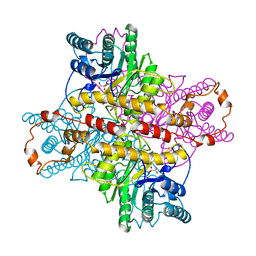 | | MECHANISM FOR ALDOSE-KETOSE INTERCONVERSION BY D-XYLOSE ISOMERASE INVOLVING RING OPENING FOLLOWED BY A 1,2-HYDRIDE SHIFT | | Descriptor: | D-XYLOSE ISOMERASE, D-gluconic acid, MANGANESE (II) ION | | Authors: | Collyer, C.A, Henrick, K, Blow, D.M. | | Deposit date: | 1991-10-09 | | Release date: | 1993-07-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanism for aldose-ketose interconversion by D-xylose isomerase involving ring opening followed by a 1,2-hydride shift.
J.Mol.Biol., 212, 1990
|
|
1XLG
 
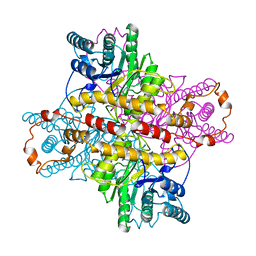 | | MECHANISM FOR ALDOSE-KETOSE INTERCONVERSION BY D-XYLOSE ISOMERASE INVOLVING RING OPENING FOLLOWED BY A 1,2-HYDRIDE SHIFT | | Descriptor: | ALUMINUM ION, D-XYLOSE ISOMERASE, MAGNESIUM ION, ... | | Authors: | Collyer, C.A, Henrick, K, Blow, D.M. | | Deposit date: | 1991-10-09 | | Release date: | 1993-07-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanism for aldose-ketose interconversion by D-xylose isomerase involving ring opening followed by a 1,2-hydride shift.
J.Mol.Biol., 212, 1990
|
|
1XLH
 
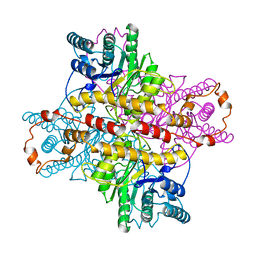 | | MECHANISM FOR ALDOSE-KETOSE INTERCONVERSION BY D-XYLOSE ISOMERASE INVOLVING RING OPENING FOLLOWED BY A 1,2-HYDRIDE SHIFT | | Descriptor: | ALUMINUM ION, D-XYLOSE ISOMERASE | | Authors: | Collyer, C.A, Henrick, K, Blow, D.M. | | Deposit date: | 1991-10-09 | | Release date: | 1993-07-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanism for aldose-ketose interconversion by D-xylose isomerase involving ring opening followed by a 1,2-hydride shift.
J.Mol.Biol., 212, 1990
|
|
1XLI
 
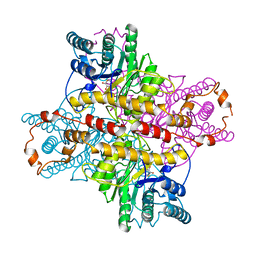 | | MECHANISM FOR ALDOSE-KETOSE INTERCONVERSION BY D-XYLOSE ISOMERASE INVOLVING RING OPENING FOLLOWED BY A 1,2-HYDRIDE SHIFT | | Descriptor: | 5-thio-alpha-D-glucopyranose, D-XYLOSE ISOMERASE, MANGANESE (II) ION | | Authors: | Collyer, C.A, Henrick, K, Blow, D.M. | | Deposit date: | 1991-10-09 | | Release date: | 1993-07-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanism for aldose-ketose interconversion by D-xylose isomerase involving ring opening followed by a 1,2-hydride shift.
J.Mol.Biol., 212, 1990
|
|
1XLJ
 
 | | MECHANISM FOR ALDOSE-KETOSE INTERCONVERSION BY D-XYLOSE ISOMERASE INVOLVING RING OPENING FOLLOWED BY A 1,2-HYDRIDE SHIFT | | Descriptor: | D-XYLOSE ISOMERASE, MANGANESE (II) ION, Xylitol | | Authors: | Collyer, C.A, Henrick, K, Blow, D.M. | | Deposit date: | 1991-10-09 | | Release date: | 1993-07-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanism for aldose-ketose interconversion by D-xylose isomerase involving ring opening followed by a 1,2-hydride shift.
J.Mol.Biol., 212, 1990
|
|
1XLK
 
 | | MECHANISM FOR ALDOSE-KETOSE INTERCONVERSION BY D-XYLOSE ISOMERASE INVOLVING RING OPENING FOLLOWED BY A 1,2-HYDRIDE SHIFT | | Descriptor: | D-XYLOSE ISOMERASE, MANGANESE (II) ION | | Authors: | Collyer, C.A, Henrick, K, Blow, D.M. | | Deposit date: | 1991-10-09 | | Release date: | 1993-07-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanism for aldose-ketose interconversion by D-xylose isomerase involving ring opening followed by a 1,2-hydride shift.
J.Mol.Biol., 212, 1990
|
|
1XLL
 
 | | MECHANISM FOR ALDOSE-KETOSE INTERCONVERSION BY D-XYLOSE ISOMERASE INVOLVING RING OPENING FOLLOWED BY A 1,2-HYDRIDE SHIFT | | Descriptor: | D-XYLOSE ISOMERASE, ZINC ION | | Authors: | Collyer, C.A, Henrick, K, Blow, D.M. | | Deposit date: | 1991-10-09 | | Release date: | 1993-07-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanism for aldose-ketose interconversion by D-xylose isomerase involving ring opening followed by a 1,2-hydride shift.
J.Mol.Biol., 212, 1990
|
|
1XLM
 
 | | D254E, D256E MUTANT OF D-XYLOSE ISOMERASE COMPLEXED WITH AL3 AND XYLITOL | | Descriptor: | ALUMINUM ION, D-XYLOSE ISOMERASE, Xylitol | | Authors: | Gerczei, T, Bocskei, Z.S, Szabo, E, Naray-Szabo, G, Asboth, B. | | Deposit date: | 1997-07-22 | | Release date: | 1998-01-28 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure determination and refinement of the Al3+ complex of the D254,256E mutant of Arthrobacter D-xylose isomerase at 2.40 A resolution. Further evidence for inhibitor-induced metal ion movement.
Int.J.Biol.Macromol., 25, 1999
|
|
1XLN
 
 | |
1XLO
 
 | |
1XLP
 
 | |
1XLQ
 
 | |
1XLR
 
 | | CHORISMATE LYASE WITH INHIBITOR VANILLATE | | Descriptor: | 4-HYDROXY-3-METHOXYBENZOATE, Chorismate--pyruvate lyase | | Authors: | Gallagher, D.T, Smith, N. | | Deposit date: | 2004-09-30 | | Release date: | 2004-11-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural analysis of ligand binding and catalysis in chorismate lyase.
Arch.Biochem.Biophys., 445, 2006
|
|
1XLS
 
 | | Crystal structure of the mouse CAR/RXR LBD heterodimer bound to TCPOBOP and 9cRA and a TIF2 peptide containg the third LXXLL motifs | | Descriptor: | (9cis)-retinoic acid, 3,5-DICHLORO-2-{4-[(3,5-DICHLOROPYRIDIN-2-YL)OXY]PHENOXY}PYRIDINE, Nuclear receptor coactivator 2, ... | | Authors: | Suino, K, peng, L, Reynolds, R, Li, Y, Cha, J.-Y, Repa, J.J, Kliewer, S.A, Xu, H.E. | | Deposit date: | 2004-09-30 | | Release date: | 2004-12-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | The nuclear xenobiotic receptor CAR: structural determinants of constitutive activation and heterodimerization.
Mol.Cell, 16, 2004
|
|
1XLT
 
 | | Crystal structure of Transhydrogenase [(domain I)2:domain III] heterotrimer complex | | Descriptor: | NAD(P) transhydrogenase subunit alpha part 1, NAD(P) transhydrogenase subunit beta, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Sundaresan, V, Chartron, J, Yamaguchi, M, Stout, C.D. | | Deposit date: | 2004-09-30 | | Release date: | 2005-04-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Conformational Diversity in NAD(H) and Transhydrogenase Nicotinamide Nucleotide Binding Domains
J.Mol.Biol., 346, 2005
|
|
1XLU
 
 | | X-Ray Structure Of Di-Isopropyl-Phosphoro-Fluoridate (Dfp) Inhibited Butyrylcholinesterase after Aging | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Nachon, F, Asojo, O.A, Borgstahl, G.E.O, Masson, P, Lockridge, O. | | Deposit date: | 2004-09-30 | | Release date: | 2005-02-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.198 Å) | | Cite: | Role of Water in Aging of Human Butyrylcholinesterase Inhibited by Echothiophate: The Crystal Structure Suggests Two Alternative Mechanisms of Aging
Biochemistry, 44, 2005
|
|
