2OS0
 
 | | Structures of actinonin bound peptide deformylases from E. faecalis and S. pyogenes | | Descriptor: | NICKEL (II) ION, Peptide deformylase, SULFATE ION | | Authors: | Kim, E.E, Kim, K.-H, Moon, J.H, Choi, K, Lee, H.K, Park, H.S. | | Deposit date: | 2007-02-05 | | Release date: | 2008-03-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structures of actinonin bound peptide deformylases from E. faecalis and S. pyogenes
To be Published
|
|
5OWO
 
 | | Human cytoplasmic Dynein N-Terminus dimerization domain at 1.8 Angstrom resolution | | Descriptor: | CALCIUM ION, Cytoplasmic dynein 1 heavy chain 1, GLYCEROL, ... | | Authors: | Urnavicius, L, Lau, C.K, Elshenawy, M.M, Morales-Rios, E, Motz, C, Yildiz, A, Carter, A.P. | | Deposit date: | 2017-09-01 | | Release date: | 2018-07-11 | | Last modified: | 2022-03-30 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Cryo-EM shows how dynactin recruits two dyneins for faster movement.
Nature, 554, 2018
|
|
2K6S
 
 | |
3EPL
 
 | |
3EPJ
 
 | |
3EPK
 
 | |
3EPH
 
 | |
2LSK
 
 | | C-terminal domain of human REV1 in complex with DNA-polymerase H (eta) | | Descriptor: | DNA polymerase eta, DNA repair protein REV1 | | Authors: | Pozhidaeva, A, Pustovalova, Y, Bezsonova, I, Korzhnev, D. | | Deposit date: | 2012-05-01 | | Release date: | 2012-06-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structure and dynamics of the C-terminal domain from human Rev1 and its complex with Rev1 interacting region of DNA polymerase eta.
Biochemistry, 51, 2012
|
|
2LQY
 
 | | Structure and orientation of the gH625-644 membrane interacting region of herpes simplex virus type 1 in a membrane mimetic system. | | Descriptor: | Envelope glycoprotein H | | Authors: | Isernia, C, Galdiero, S, Russo, L, Falanga, A, Cantisani, M, Vitiello, M, Fattorusso, R, Malgieri, G, Galdiero, M. | | Deposit date: | 2012-03-19 | | Release date: | 2012-04-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure and Orientation of the gH625-644 Membrane Interacting Region of Herpes Simplex Virus Type 1 in a Membrane Mimetic System.
Biochemistry, 51, 2012
|
|
1O9F
 
 | | Structural view of a fungal toxin acting on a 14-3-3 regulatory complex | | Descriptor: | 14-3-3-LIKE PROTEIN C, FUSICOCCIN, PLASMA MEMBRANE H+ ATPASE | | Authors: | Wurtele, M, Jelich-Ottmann, C, Wittinghofer, A, Oecking, C. | | Deposit date: | 2002-12-12 | | Release date: | 2003-03-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural View of a Fungal Toxin Acting on a 14-3-3 Regulatory Complex
Embo J., 22, 2003
|
|
6IB6
 
 | | Solution structure of the water-soluble LU-domain of human Lypd6 protein | | Descriptor: | Ly6/PLAUR domain-containing protein 6 | | Authors: | Tsarev, A.V, Kulbatskii, D.S, Paramonov, A.S, Lyukmanova, E.N, Shenkarev, Z.O. | | Deposit date: | 2018-11-29 | | Release date: | 2019-12-18 | | Last modified: | 2021-01-13 | | Method: | SOLUTION NMR | | Cite: | Structural Diversity and Dynamics of Human Three-Finger Proteins Acting on Nicotinic Acetylcholine Receptors.
Int J Mol Sci, 21, 2020
|
|
6SN6
 
 | |
1O9D
 
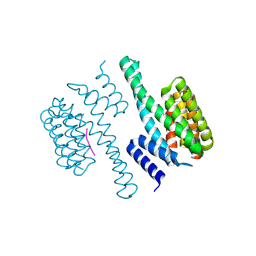 | | Structural view of a fungal toxin acting on a 14-3-3 regulatory complex | | Descriptor: | 14-3-3-LIKE PROTEIN C, PLASMA MEMBRANE H+ ATPASE | | Authors: | Wurtele, M, Jelich-Ottmann, C, Wittinghofer, A, Oecking, C. | | Deposit date: | 2002-12-12 | | Release date: | 2003-03-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural View of a Fungal Toxin Acting on a 14-3-3 Regulatory Complex
Embo J., 22, 2003
|
|
6T9Y
 
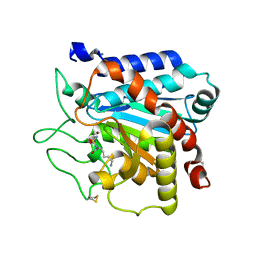 | |
6TNK
 
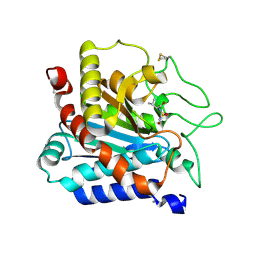 | |
6GO2
 
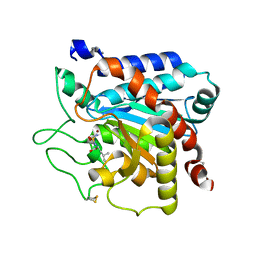 | |
1O9C
 
 | | Structural view of a fungal toxin acting on a 14-3-3 regulatory complex | | Descriptor: | 14-3-3-LIKE PROTEIN C, CHLORIDE ION, CITRATE ANION | | Authors: | Wurtele, M, Jelich-Ottmann, C, Wittinghofer, A, Oecking, C. | | Deposit date: | 2002-12-12 | | Release date: | 2003-03-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural View of a Fungal Toxin Acting on a 14-3-3 Regulatory Complex
Embo J., 22, 2003
|
|
1O9E
 
 | | Structural view of a fungal toxin acting on a 14-3-3 regulatory complex | | Descriptor: | 14-3-3-LIKE PROTEIN C, CITRATE ANION, FUSICOCCIN | | Authors: | Wurtele, M, Jelich-Ottmann, C, Wittinghofer, A, Oecking, C. | | Deposit date: | 2002-12-12 | | Release date: | 2003-03-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural View of a Fungal Toxin Acting on a 14-3-3 Regulatory Complex
Embo J., 22, 2003
|
|
3OO6
 
 | |
3OO8
 
 | |
3OOA
 
 | |
3OO7
 
 | |
3OO9
 
 | |
7LMS
 
 | |
6TDA
 
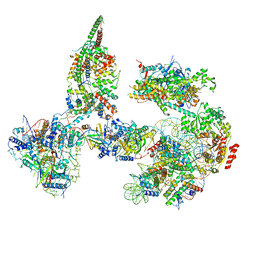 | | Structure of SWI/SNF chromatin remodeler RSC bound to a nucleosome | | Descriptor: | Actin-like protein ARP9, Actin-related protein 7, Chromatin structure-remodeling complex protein RSC58, ... | | Authors: | Wagner, F.R, Dienemann, C, Wang, H, Stuetzer, A, Tegunov, D, Urlaub, H, Cramer, P. | | Deposit date: | 2019-11-08 | | Release date: | 2020-03-18 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (15 Å) | | Cite: | Structure of SWI/SNF chromatin remodeller RSC bound to a nucleosome.
Nature, 579, 2020
|
|
