1KME
 
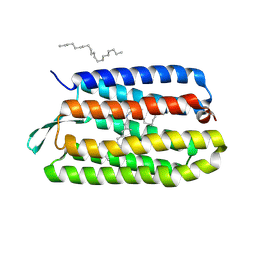 | | CRYSTAL STRUCTURE OF BACTERIORHODOPSIN CRYSTALLIZED FROM BICELLES | | Descriptor: | 2,10,23-TRIMETHYL-TETRACOSANE, Bacteriorhodopsin, RETINAL, ... | | Authors: | Faham, S, Bowie, J.U. | | Deposit date: | 2001-12-14 | | Release date: | 2002-02-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Bicelle crystallization: a new method for crystallizing membrane proteins yields a monomeric bacteriorhodopsin structure.
J.Mol.Biol., 316, 2002
|
|
1KMV
 
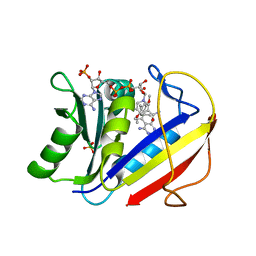 | | HUMAN DIHYDROFOLATE REDUCTASE COMPLEXED WITH NADPH AND (Z)-6-(2-[2,5-DIMETHOXYPHENYL]ETHEN-1-YL)-2,4-DIAMINO-5-METHYLPYRIDO[2,3-D]PYRIMIDINE (SRI-9662), A LIPOPHILIC ANTIFOLATE | | Descriptor: | (Z)-6-(2-[2,5-DIMETHOXYPHENYL]ETHEN-1-YL)-2,4-DIAMINO-5-METHYLPYRIDO[2,3-D]PYRIMIDINE, DIHYDROFOLATE REDUCTASE, DIMETHYL SULFOXIDE, ... | | Authors: | Klon, A.E, Heroux, A, Ross, L.J, Pathak, V, Johnson, C.A, Piper, J.R, Borhani, D.W. | | Deposit date: | 2001-12-17 | | Release date: | 2002-07-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Atomic structures of human dihydrofolate reductase complexed with NADPH and two lipophilic antifolates at 1.09 a and 1.05 a resolution.
J.Mol.Biol., 320, 2002
|
|
1KAX
 
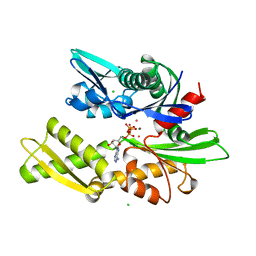 | | 70KD HEAT SHOCK COGNATE PROTEIN ATPASE DOMAIN, K71M MUTANT | | Descriptor: | 70KD HEAT SHOCK COGNATE PROTEIN, ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, ... | | Authors: | O'Brien, M.C, Flaherty, K.M, Mckay, D.B. | | Deposit date: | 1996-04-15 | | Release date: | 1996-11-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Lysine 71 of the chaperone protein Hsc70 Is essential for ATP hydrolysis.
J.Biol.Chem., 271, 1996
|
|
1KAZ
 
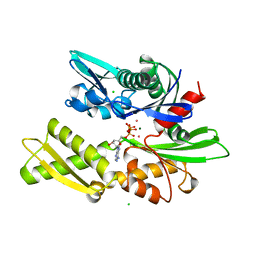 | | 70KD HEAT SHOCK COGNATE PROTEIN ATPASE DOMAIN, K71E MUTANT | | Descriptor: | 70KD HEAT SHOCK COGNATE PROTEIN, ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, ... | | Authors: | O'Brien, M.C, Flaherty, K.M, Mckay, D.B. | | Deposit date: | 1996-04-15 | | Release date: | 1996-11-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Lysine 71 of the chaperone protein Hsc70 Is essential for ATP hydrolysis.
J.Biol.Chem., 271, 1996
|
|
1L77
 
 | |
1L80
 
 | |
1L40
 
 | |
1I7Q
 
 | | ANTHRANILATE SYNTHASE FROM S. MARCESCENS | | Descriptor: | ANTHRANILATE SYNTHASE, BENZOIC ACID, GLUTAMIC ACID, ... | | Authors: | Spraggon, G, Kim, C, Nguyen-Huu, X, Yee, M.-C, Yanofsky, C, Mills, S.E. | | Deposit date: | 2001-03-10 | | Release date: | 2001-05-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The structures of anthranilate synthase of Serratia marcescens crystallized in the presence of (i) its substrates, chorismate and glutamine, and a product, glutamate, and (ii) its end-product inhibitor, L-tryptophan.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1I7P
 
 | |
1I7S
 
 | | ANTHRANILATE SYNTHASE FROM SERRATIA MARCESCENS IN COMPLEX WITH ITS END PRODUCT INHIBITOR L-TRYPTOPHAN | | Descriptor: | ANTHRANILATE SYNTHASE, TRPG, TRYPTOPHAN | | Authors: | Spraggon, G, Kim, C, Nguyen-Huu, X, Yee, M.-C, Yanofsky, C, Mills, S.E. | | Deposit date: | 2001-03-10 | | Release date: | 2001-05-16 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The structures of anthranilate synthase of Serratia marcescens crystallized in the presence of (i) its substrates, chorismate and glutamine, and a product, glutamate, and (ii) its end-product inhibitor, L-tryptophan.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1L38
 
 | |
1L79
 
 | |
5D9I
 
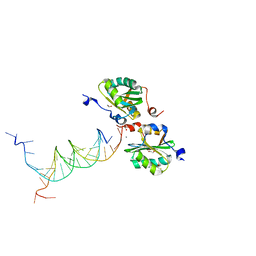 | | SV40 Large T antigen origin binding domain bound to artificial DNA fork | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Meinke, G, Bohm, A, Bullock, P.A. | | Deposit date: | 2015-08-18 | | Release date: | 2015-09-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-based analysis of the interaction between the simian virus 40 T-antigen origin binding domain and single-stranded DNA.
J. Virol., 85, 2011
|
|
8Q3E
 
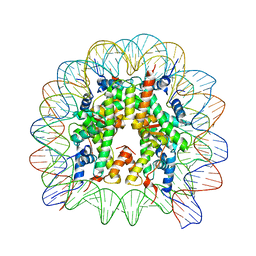 | | High Resolution Structure of Nucleosome Core with Bound Foamy Virus GAG Peptide | | Descriptor: | DNA (145-MER), GLY-GLY-TYR-ASN-LEU-ARG-PRO-ARG-THR-TYR-GLN-PRO-GLN-ARG-TYR-GLY-GLY-GLY, Histone H2A type 1-B/E, ... | | Authors: | De Falco, L, Batchelor, L.K, Dyson, P.J, Davey, C.A. | | Deposit date: | 2023-08-04 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.174 Å) | | Cite: | Viral peptide conjugates for metal-warhead delivery to chromatin.
Rsc Adv, 14, 2024
|
|
1L41
 
 | |
1L37
 
 | |
1L5T
 
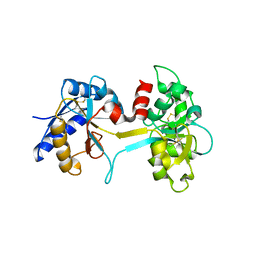 | | Crystal Structure of a Domain-Opened Mutant (R121D) of the Human Lactoferrin N-lobe Refined From a Merohedrally-Twinned Crystal Form. | | Descriptor: | lactoferrin | | Authors: | Jameson, G.B, Anderson, B.F, Breyer, W.A, Tweedie, J.W, Baker, E.N. | | Deposit date: | 2002-03-07 | | Release date: | 2002-03-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of a domain-opened mutant (R121D) of the human lactoferrin N-lobe refined from a merohedrally twinned crystal form.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1L60
 
 | |
1L56
 
 | |
1ZBB
 
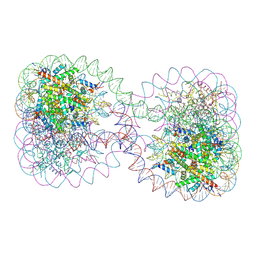 | | Structure of the 4_601_167 Tetranucleosome | | Descriptor: | DNA STRAND 1 (ARBITRARY MODEL SEQUENCE), DNA STRAND 2 (ARBITRARY MODEL SEQUENCE), HISTONE H3, ... | | Authors: | Schalch, T, Duda, S, Sargent, D.F, Richmond, T.J. | | Deposit date: | 2005-04-08 | | Release date: | 2005-07-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (9 Å) | | Cite: | X-ray structure of a tetranucleosome and its implications for the chromatin fibre.
Nature, 436, 2005
|
|
1KH4
 
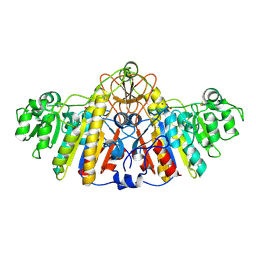 | | E. COLI ALKALINE PHOSPHATASE MUTANT (D330N) IN COMPLEX WITH PHOSPHATE | | Descriptor: | ALKALINE PHOSPHATASE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Le Du, M.H, Lamoure, C, Muller, B.H, Bulgakov, O.V, Lajeunesse, E. | | Deposit date: | 2001-11-29 | | Release date: | 2002-03-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Artificial evolution of an enzyme active site: structural studies of three highly active mutants of Escherichia coli alkaline phosphatase.
J.Mol.Biol., 316, 2002
|
|
1KH5
 
 | | E. COLI ALKALINE PHOSPHATASE MUTANT (D330N) MIMIC OF THE TRANSITION STATES WITH ALUMINIUM FLUORIDE | | Descriptor: | ALKALINE PHOSPHATASE, ALUMINUM FLUORIDE, MAGNESIUM ION, ... | | Authors: | Le Du, M.H, Lamoure, C, Muller, B.H, Bulgakov, O.V, Lajeunesse, E. | | Deposit date: | 2001-11-29 | | Release date: | 2002-03-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Artificial evolution of an enzyme active site: structural studies of three highly active mutants of Escherichia coli alkaline phosphatase.
J.Mol.Biol., 316, 2002
|
|
1KHJ
 
 | | E. COLI ALKALINE PHOSPHATASE MUTANT (D153HD330N) MIMIC OF THE TRANSITION STATES WITH ALUMINIUM FLUORIDE | | Descriptor: | ALUMINUM FLUORIDE, Alkaline phosphatase, ZINC ION | | Authors: | Le Du, M.H, Lamoure, C, Muller, B.H, Bulgakov, O.V, Lajeunesse, E, Menez, A, Boulain, J.C. | | Deposit date: | 2001-11-30 | | Release date: | 2002-03-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Artificial evolution of an enzyme active site: structural studies of three highly active mutants of Escherichia coli alkaline phosphatase.
J.Mol.Biol., 316, 2002
|
|
1L82
 
 | |
1LRL
 
 | | Crystal Structure of UDP-Galactose 4-Epimerase Mutant Y299C Complexed with UDP-Glucose | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SODIUM ION, TRIETHYLENE GLYCOL, ... | | Authors: | Thoden, J.B, Henderson, J.M, Fridovich-Keil, J.L, Holden, H.M. | | Deposit date: | 2002-05-15 | | Release date: | 2002-07-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural analysis of the Y299C mutant of Escherichia coli UDP-galactose 4-epimerase. Teaching an old dog new tricks.
J.Biol.Chem., 277, 2002
|
|
