9CJ7
 
 | | Lineage IV Lassa virus glycoprotein (Josiah) in complex with monoclonal antibody 8.9F | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Brouwer, P.J.M, Perrett, H.R, Ward, A.B. | | Deposit date: | 2024-07-05 | | Release date: | 2024-09-25 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Defining bottlenecks and opportunities for Lassa virus neutralization by structural profiling of vaccine-induced polyclonal antibody responses.
Cell Rep, 43, 2024
|
|
8DJS
 
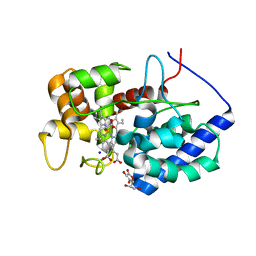 | |
8YMZ
 
 | | Structure of ZBTB43 in complex with CACA containing B-form DNA | | Descriptor: | DNA (5'-D(*AP*AP*GP*TP*GP*TP*GP*TP*AP*TP*GP*TP*GP*TP*G)-3'), DNA (5'-D(*TP*CP*AP*CP*AP*CP*AP*TP*AP*CP*AP*CP*AP*CP*T)-3'), ZINC ION, ... | | Authors: | Li, X, Yang, Y. | | Deposit date: | 2024-03-10 | | Release date: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural insights into the recognition of purine-pyrimidine dinucleotide repeats by zinc finger protein ZBTB43.
Febs J., 2024
|
|
9ERX
 
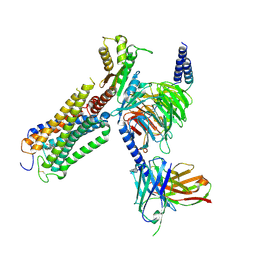 | | Structural basis of D9-THC analog activity at the Cannabinoid 1 receptor | | Descriptor: | (6aR,10aR)-9-(hydroxymethyl)-6,6-dimethyl-3-(2-methyloctan-2-yl)-6a,7,10,10a-tetrahydrobenzo[c]chromen-1-ol, Antibody ScFv16 Fab fragment, Cannabinoid receptor 1, ... | | Authors: | Thorsen, T.S, Kulkarni, Y, Boggild, A, Drace, T, Nissen, P, Gajhede, M, Boesen, T, Kastrup, J.S, Gloriam, D. | | Deposit date: | 2024-03-25 | | Release date: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of Delta 9 -THC analog activity at the Cannabinoid 1 receptor.
Res Sq, 2024
|
|
9EYU
 
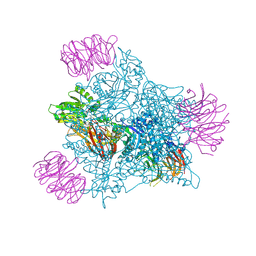 | | Human PRMT5 in complex with AZ compound 1 | | Descriptor: | (1~{S})-2-[(2-carbamimidamido-1,3-thiazol-5-yl)methyl]-~{N}-[(4-fluorophenyl)methyl]-3-oxidanylidene-1~{H}-isoindole-1-carboxamide, 5'-DEOXY-5'-METHYLTHIOADENOSINE, Methylosome protein WDR77, ... | | Authors: | Debreczeni, J. | | Deposit date: | 2024-04-09 | | Release date: | 2024-08-14 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Discovery and In Vivo Efficacy of AZ-PRMT5i-1, a Novel PRMT5 Inhibitor with High MTA Cooperativity.
J.Med.Chem., 67, 2024
|
|
9CX6
 
 | |
9FP0
 
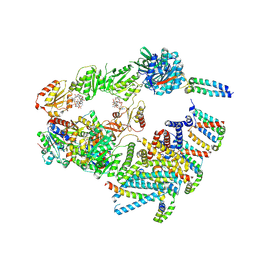 | | Cryo-EM structure of the 'crown'less Bcs macrocomplex for E. coli cellulose secretion in non-saturating c-di-GMP (local) | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), ADENOSINE-5'-TRIPHOSPHATE, Cell division protein, ... | | Authors: | Anso, I, Krasteva, P.V. | | Deposit date: | 2024-06-12 | | Release date: | 2024-10-16 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Structural basis for synthase activation and cellulose modification in the E. coli Type II Bcs secretion system.
Nat Commun, 15, 2024
|
|
8YN4
 
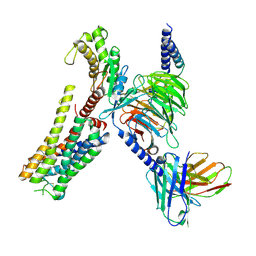 | | Cryo-EM structure of histamine H2 receptor in complex with histamine and miniGq | | Descriptor: | Antibody fragment scFv16, CHOLESTEROL, Engineered guanine nucleotide-binding protein G(q) subunit alpha, ... | | Authors: | Zhang, X, Liu, G, Li, X, Gong, W. | | Deposit date: | 2024-03-10 | | Release date: | 2024-10-09 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Structural basis of ligand recognition and activation of the histamine receptor family.
Nat Commun, 15, 2024
|
|
9F1B
 
 | | Mammalian ternary complex of a translating 80S ribosome, NAC and NatA/E | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S11, ... | | Authors: | Yudin, D, Scaiola, A, Ban, N. | | Deposit date: | 2024-04-18 | | Release date: | 2024-08-21 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | NAC guides a ribosomal multienzyme complex for nascent protein processing.
Nature, 633, 2024
|
|
9IJE
 
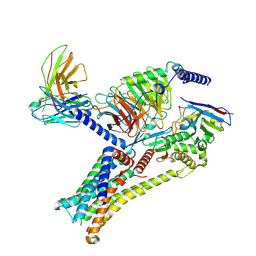 | | Epinephrine-activated human beta3 adrenergic receptor | | Descriptor: | Beta-3 adrenergic receptor, Camelid antibody VHH fragment, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Zheng, S, Zhang, S, Dai, S, Chen, K, Gao, K, Lin, B, Liu, X. | | Deposit date: | 2024-06-22 | | Release date: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (2.34 Å) | | Cite: | Molecular Mechanism of the beta 3AR Agonist Activity of a beta-Blocker.
Chempluschem, 2024
|
|
8YN3
 
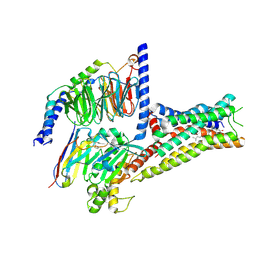 | | Cryo-EM structure of histamine H2 receptor in complex with histamine and miniGs | | Descriptor: | CHOLESTEROL, Engineered guanine nucleotide,binding protein G(s) subunit alpha, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Zhang, X, Liu, G, Li, X, Gong, W. | | Deposit date: | 2024-03-10 | | Release date: | 2024-10-09 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.56 Å) | | Cite: | Structural basis of ligand recognition and activation of the histamine receptor family.
Nat Commun, 15, 2024
|
|
9G5L
 
 | |
9FIJ
 
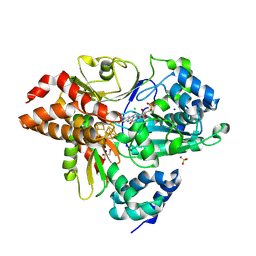 | | Crystal Structure of reduced NuoEF variant E222K(NuoF) from Aquifex aeolicus | | Descriptor: | 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, CHLORIDE ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Wohlwend, D, Friedrich, T, Goeppert-Asadollahpour, S. | | Deposit date: | 2024-05-29 | | Release date: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural robustness of the NADH binding site in NADH:ubiquinone oxidoreductase (complex I).
Biochim Biophys Acta Bioenerg, 1865, 2024
|
|
8DJU
 
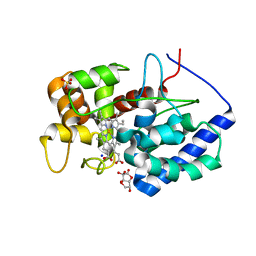 | | Cytosolic ascorbate peroxidase from Sorghum bicolor - bicyclic dehydroascorbic acid complex | | Descriptor: | (3aS,6S,6aR)-3,3,3a,6-tetrahydroxytetrahydrofuro[3,2-b]furan-2(3H)-one (non-preferred name), L-ascorbate peroxidase, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Zhang, B, Kang, C. | | Deposit date: | 2022-07-01 | | Release date: | 2023-01-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A sorghum ascorbate peroxidase with four binding sites has activity against ascorbate and phenylpropanoids.
Plant Physiol., 192, 2023
|
|
9CEX
 
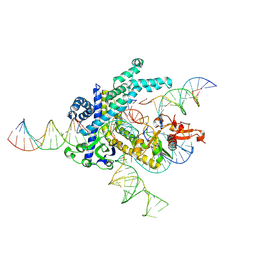 | | Spizellomyces punctatus Fanzor (SpuFz) State 4 | | Descriptor: | DNA (29-MER), DNA (5'-D(*(MG)*(MG)P*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*A)-3'), DNA (5'-D(P*CP*GP*GP*TP*AP*CP*CP*CP*GP*GP*GP*CP*AP*TP*A)-3'), ... | | Authors: | Xu, P, Saito, M, Zhang, F. | | Deposit date: | 2024-06-27 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Structural insights into the diversity and DNA cleavage mechanism of Fanzor.
Cell, 187, 2024
|
|
9BF5
 
 | |
9BPF
 
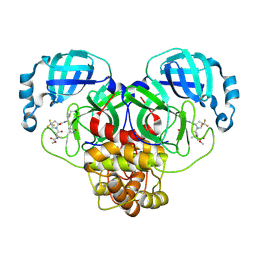 | | Crystal structure of main protease of SARS-CoV-2 complexed with inhibitor | | Descriptor: | 3C-like proteinase nsp5, N-(methoxycarbonyl)-3-methyl-L-valyl-(4R)-N-{(1Z,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-4-(trifluoromethyl)-L-prolinamide | | Authors: | Chen, P, Arutyunova, E, Lemieux, M.J. | | Deposit date: | 2024-05-07 | | Release date: | 2024-08-07 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Structural Comparison of Oral SARS-CoV-2 Drug Candidate Ibuzatrelvir Complexed with the Main Protease (M pro ) of SARS-CoV-2 and MERS-CoV.
Jacs Au, 4, 2024
|
|
9BEW
 
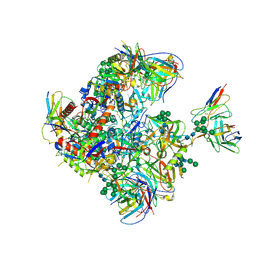 | |
9IXN
 
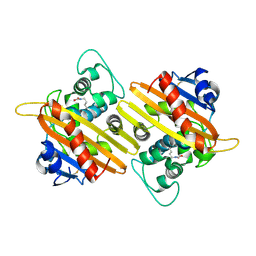 | | Crystal structure of OXA-10 | | Descriptor: | Beta-lactamase OXA-10 | | Authors: | Lee, C.E, Park, Y.S, Park, H.J, Kang, L.W. | | Deposit date: | 2024-07-29 | | Release date: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural Insights into Alterations in the Substrate Spectrum of Serine-beta-Lactamase OXA-10 from Pseudomonas aeruginosa by Single Amino Acid Substitutions.
Emerg Microbes Infect, 2024
|
|
9CJ0
 
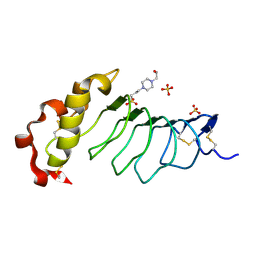 | |
9G1Y
 
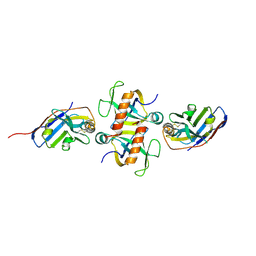 | |
9B9Q
 
 | |
9ENN
 
 | | L-amino acid oxidase 4 (HcLAAO4) from the fungus Hebeloma cylindrosporum in complex with N-epsilon-acetyl-L-lysine | | Descriptor: | DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, L-amino acid oxidase 4, N(6)-ACETYLLYSINE, ... | | Authors: | Gilzer, D, Koopmeiners, S, Fischer von Mollard, G, Niemann, H.H. | | Deposit date: | 2024-03-13 | | Release date: | 2024-08-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure and enzyme engineering of the broad substrate spectrum l-amino acid oxidase 4 from the fungus Hebeloma cylindrosporum.
Febs Lett., 598, 2024
|
|
9BOK
 
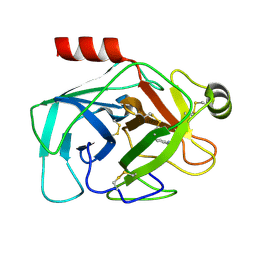 | | Crystal structure of reduced bovine trypsin, 50mM DTT-treated | | Descriptor: | BENZAMIDINE, Cationic trypsin | | Authors: | Zhou, D, Chen, L, Rose, J.P, Wang, B.C. | | Deposit date: | 2024-05-03 | | Release date: | 2024-08-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal structure of reduced bovine trypsin, 50mM DTT-treated
To Be Published
|
|
9G2G
 
 | | Staphylococcus aureus MazF in complex with nanobody 5 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Endoribonuclease MazF, Nanobody 5 | | Authors: | Prolic-Kalinsek, M, Zorzini, V, Haesaerts, S, Loris, R. | | Deposit date: | 2024-07-10 | | Release date: | 2024-07-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.80048239 Å) | | Cite: | Nanobody-mediated activation and inhibition of Staphylococcus aureus MazF
To Be Published
|
|
