7BG5
 
 | |
7B8Z
 
 | | Notum-Fragment 277 | | Descriptor: | 2-[4-(3-chlorophenyl)piperazin-1-ium-1-yl]ethanenitrile, 2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, ... | | Authors: | Zhao, Y, Jonees, E.Y. | | Deposit date: | 2020-12-13 | | Release date: | 2022-01-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structural Analysis and Development of Notum Fragment Screening Hits.
Acs Chem Neurosci, 13, 2022
|
|
7F1X
 
 | | X-ray crystal structure of visual arrestin complexed with inositol 1,4,5-triphosphate | | Descriptor: | 1,2-ETHANEDIOL, D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, PENTANEDIAL, ... | | Authors: | Jang, K, Kang, M, Eger, B.T, Choe, H.W, Ernst, O.P, Kim, Y.J. | | Deposit date: | 2021-06-10 | | Release date: | 2021-10-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural evidence for visual arrestin priming via complexation of phosphoinositols.
Structure, 30, 2022
|
|
9INP
 
 | | Human Pin1 (Peptidyl-prolyl cis-trans isomerase) catalytic domain in complex with a covalent inhibitor | | Descriptor: | CITRIC ACID, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1, ~{N}-[(3~{R})-1,1-bis(oxidanylidene)thiolan-3-yl]-2-chloranyl-~{N}-(2,2-dimethylpropyl)-5-nitro-pyrimidin-4-amine | | Authors: | Wang, X.Y, Tian, M.Z, Zhou, J, Xu, B.L. | | Deposit date: | 2024-07-08 | | Release date: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Discovery of Novel Pyrimidine Derivatives as Human Pin1 Covalent Inhibitors.
Acs Med.Chem.Lett., 16, 2025
|
|
6TXO
 
 | | Crystal structure of the haemagglutinin mutant (Gln226Leu, Del228) from an H10N7 seal influenza virus isolated in Germany in complex with avian receptor analogue 3'-SLN | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Xiong, X, Purkiss, A, Walker, P, Gamblin, S, Skehel, J.J. | | Deposit date: | 2020-01-14 | | Release date: | 2020-10-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Hemagglutinin Traits Determine Transmission of Avian A/H10N7 Influenza Virus between Mammals.
Cell Host Microbe, 28, 2020
|
|
6EV7
 
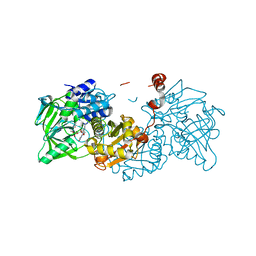 | | Structure of E282D A. niger Fdc1 with prFMN in the iminium form | | Descriptor: | 1-deoxy-5-O-phosphono-1-(3,3,4,5-tetramethyl-9,11-dioxo-2,3,8,9,10,11-hexahydro-7H-quinolino[1,8-fg]pteridin-12-ium-7-y l)-D-ribitol, Ferulic acid decarboxylase 1, MANGANESE (II) ION, ... | | Authors: | Bailey, S.S, Leys, D, Payne, K.A.P. | | Deposit date: | 2017-11-01 | | Release date: | 2017-12-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | The role of conserved residues in Fdc decarboxylase in prenylated flavin mononucleotide oxidative maturation, cofactor isomerization, and catalysis.
J. Biol. Chem., 293, 2018
|
|
7V7B
 
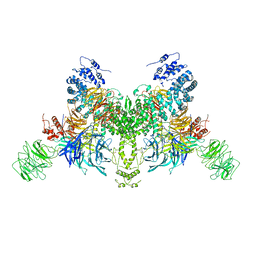 | |
7VC3
 
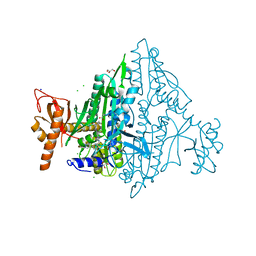 | | Toxoplasma gondii Prolyl-tRNA Synthetase (TgPRS) in Complex with inhibitor L97 and L-proline at 1.97 A resolution | | Descriptor: | 4-[(3S)-3-cyclopropyl-3-(hydroxymethyl)-2-oxidanylidene-pyrrolidin-1-yl]-N-[[3-fluoranyl-5-(1-methylpyrazol-4-yl)phenyl]methyl]-6-methyl-pyridine-2-carboxamide, CHLORIDE ION, ETHANOLAMINE, ... | | Authors: | Malhotra, N, Yogavel, M, Sharma, A. | | Deposit date: | 2021-09-01 | | Release date: | 2022-09-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.973 Å) | | Cite: | Targeting prolyl-tRNA synthetase via a series of ATP-mimetics to accelerate drug discovery against toxoplasmosis.
Plos Pathog., 19, 2023
|
|
4ZT5
 
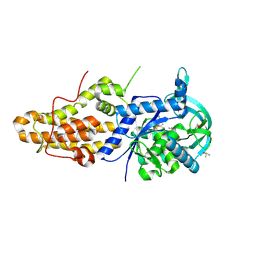 | | Trypanosoma brucei methionyl-tRNA synthetase in complex with inhibitor (2S)-N-(3,5-dichlorobenzyl)-N'-(1H-imidazo[4,5-b]pyridin-2-yl)-2-methylpropane-1,3-diamine (Chem 1655) | | Descriptor: | (2S)-N-(3,5-dichlorobenzyl)-N'-(1H-imidazo[4,5-b]pyridin-2-yl)-2-methylpropane-1,3-diamine, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Koh, C.-Y, Hol, W.G.J. | | Deposit date: | 2015-05-14 | | Release date: | 2016-05-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | 5-Fluoroimidazo[4,5-b]pyridine Is a Privileged Fragment That Conveys Bioavailability to Potent Trypanosomal Methionyl-tRNA Synthetase Inhibitors.
Acs Infect Dis., 2, 2016
|
|
7UK1
 
 | |
7BO1
 
 | |
6ET0
 
 | | Crystal structure of PqsBC (C129A) mutant from Pseudomonas aeruginosa (crystal form 1) | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, ... | | Authors: | Witzgall, F, Blankenfeldt, W. | | Deposit date: | 2017-10-25 | | Release date: | 2018-07-04 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | The Alkylquinolone Repertoire of Pseudomonas aeruginosa is Linked to Structural Flexibility of the FabH-like 2-Heptyl-3-hydroxy-4(1H)-quinolone (PQS) Biosynthesis Enzyme PqsBC.
Chembiochem, 19, 2018
|
|
5Y25
 
 | | EGFR kinase domain mutant (T790M/L858R) with covalent ligand NS-062 | | Descriptor: | (2R)-N-[4-[(3-chloranyl-4-fluoranyl-phenyl)amino]-7-(3-morpholin-4-ylpropoxy)quinazolin-6-yl]-1-(2-fluoranylethanoyl)pyrrolidine-2-carboxamide, Epidermal growth factor receptor | | Authors: | Shiroishi, M, Abe, Y, Caaveiro, J.M.M, Sakamoto, S, Morimoto, S, Fuchida, H, Shindo, N, Ojida, A. | | Deposit date: | 2017-07-24 | | Release date: | 2018-07-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.102 Å) | | Cite: | Selective and reversible modification of kinase cysteines with chlorofluoroacetamides.
Nat.Chem.Biol., 15, 2019
|
|
3IUK
 
 | | Crystal structure of putative bacterial protein of unknown function (DUF885, PF05960.1, ) from Arthrobacter aurescens TC1, reveals fold similar to that of M32 carboxypeptidases | | Descriptor: | GLYCEROL, MAGNESIUM ION, uncharacterized protein | | Authors: | Nocek, B, Chhor, G, Cobb, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-08-31 | | Release date: | 2009-10-27 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of putative bacterial protein of unknown function (DUF885, PF05960.1, ) from Arthrobacter aurescens TC1, reveals fold similar to that of M32 carboxypeptidases
To be Published
|
|
5ALI
 
 | | ligand complex structure of soluble epoxide hydrolase | | Descriptor: | 1-[3-[5-(7-aminothiazolo[5,4-d]pyrimidin-2-yl)-1-[[(2S)-tetrahydrofuran-2-yl]methyl]imidazol-4-yl]phenyl]-3-(4-methoxyphenyl)urea, BIFUNCTIONAL EPOXIDE HYDROLASE 2, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Oster, L, Tapani, S, Xue, Y, Kack, H. | | Deposit date: | 2015-03-08 | | Release date: | 2015-05-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Successful Generation of Structural Information for Fragment-Based Drug Discovery.
Drug Discov Today, 20, 2015
|
|
3IHD
 
 | |
8DIS
 
 | |
5A8R
 
 | |
5KZ8
 
 | | Mark2 complex with 7-[(1S)-1-(4-fluorophenyl)ethyl]-5,5-dimethyl-2-(3-pyridylamino)pyrrolo[2,3-d]pyrimidin-6-one | | Descriptor: | 5,5-dimethyl-7-[(1~{S})-4-oxidanyl-1~{H}-inden-1-yl]-2-phenylazanyl-pyrrolo[2,3-d]pyrimidin-6-one, Serine/threonine-protein kinase MARK2 | | Authors: | Su, H.P, Munshi, S.K. | | Deposit date: | 2016-07-23 | | Release date: | 2017-05-31 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | Structure guided design of a series of selective pyrrolopyrimidinone MARK inhibitors.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
7F6S
 
 | |
7RMD
 
 | | Crystal structure of the first bromodomain of human BRD4 in complex with SJ001011461-1 | | Descriptor: | 1,2-ETHANEDIOL, 2-[(6S,10R)-4-(4-chlorophenyl)-2,3,9-trimethyl-6H-thieno[3,2-f][1,2,4]triazolo[4,3-a][1,4]diazepin-6-yl]-N-(1,3,4-thiadiazol-2-yl)acetamide, Bromodomain-containing protein 4 | | Authors: | Stachowski, T.R, Fischer, M. | | Deposit date: | 2021-07-27 | | Release date: | 2022-08-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | From PROTAC to inhibitor: Structure-guided discovery of potent and orally bioavailable BET inhibitors.
Eur.J.Med.Chem., 251, 2023
|
|
5HGH
 
 | | HLA*A2402 complexed with HIV nef138 10mer epitope | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-24 alpha chain, ... | | Authors: | Shi, Y, Qi, J, Gao, G.F. | | Deposit date: | 2016-01-08 | | Release date: | 2016-06-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.392 Å) | | Cite: | Effects of a Single Escape Mutation on T Cell and HIV-1 Co-adaptation.
Cell Rep, 15, 2016
|
|
3I7V
 
 | | Crystal structure of AP4A hydrolase complexed with AP4A (ATP) (aq_158) from Aquifex aeolicus Vf5 | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, AP4A hydrolase, ... | | Authors: | Jeyakanthan, J, Kanaujia, S.P, Nakagawa, N, Sekar, K, Kuramitsu, S, Shinkai, A, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-07-09 | | Release date: | 2009-07-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Free and ATP-bound structures of Ap(4)A hydrolase from Aquifex aeolicus V5
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3I93
 
 | | Crystal structure of Mycobacterium tuberculosis dUTPase STOP138T mutant | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Deoxyuridine 5'-triphosphate nucleotidohydrolase, ... | | Authors: | Leveles, I, Harmat, V, Lopata, A, Toth, J, Vertessy, B.G. | | Deposit date: | 2009-07-10 | | Release date: | 2009-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Direct contacts between conserved motifs of different subunits provide major contribution to active site organization in human and mycobacterial dUTPases.
Febs Lett., 584, 2010
|
|
7F6F
 
 | | Crystal structure of metal-citrate-binding protein (MctA) of ABC transporter endogenously bound to Mg2+-citrate complex (Form II) | | Descriptor: | 1,2-ETHANEDIOL, CARBON DIOXIDE, CITRIC ACID, ... | | Authors: | Kanaujia, S.P, Mandal, S.K, Gogoi, P. | | Deposit date: | 2021-06-25 | | Release date: | 2021-12-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and thermodynamic insights into a novel Mg 2+ -citrate-binding protein from the ABC transporter superfamily.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
