4K5U
 
 | | Recognition of the BG-H Antigen by a Lamprey Variable Lymphocyte Receptor | | Descriptor: | Variable lymphocyte receptor, beta-D-galactopyranose | | Authors: | Luo, M, Velikovsky, C.A, Yang, X.B, Mariuzza, R.A. | | Deposit date: | 2013-04-15 | | Release date: | 2013-06-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.698 Å) | | Cite: | Recognition of the thomsen-friedenreich pancarcinoma carbohydrate antigen by a lamprey variable lymphocyte receptor.
J.Biol.Chem., 288, 2013
|
|
2QBO
 
 | | Crystal structure of the P450cam G248V mutant in the cyanide bound state | | Descriptor: | CAMPHOR, CYANIDE ION, Cytochrome P450-cam, ... | | Authors: | von Koenig, K, Makris, T.M, Sligar, S.D, Schlichting, I. | | Deposit date: | 2007-06-18 | | Release date: | 2007-12-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Alteration of P450 Distal Pocket Solvent Leads to Impaired Proton Delivery and Changes in Heme Geometry.
Biochemistry, 46, 2007
|
|
4KR4
 
 | |
1TRN
 
 | |
3HJH
 
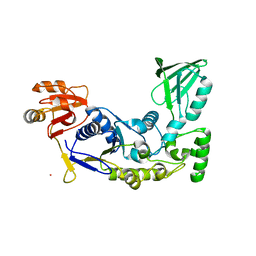 | | A rigid N-terminal clamp restrains the motor domains of the bacterial transcription-repair coupling factor | | Descriptor: | COBALT (II) ION, Transcription-repair-coupling factor | | Authors: | Murphy, M, Gong, P, Ralto, K, Manelyte, L, Savery, N, Theis, K. | | Deposit date: | 2009-05-21 | | Release date: | 2009-10-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | An N-terminal clamp restrains the motor domains of the bacterial transcription-repair coupling factor Mfd.
Nucleic Acids Res., 37, 2009
|
|
4M6E
 
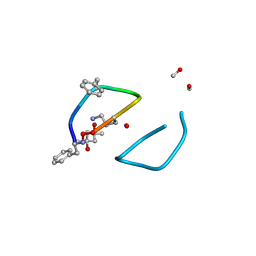 | | The high resolution structure of tyrocidine A reveals an amphipathic dimer | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, METHANOL, tyrocidine A | | Authors: | Loll, P.J, Economou, N.J, Nahoum, V. | | Deposit date: | 2013-08-09 | | Release date: | 2014-03-19 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | The high resolution structure of tyrocidine A reveals an amphipathic dimer.
Biochim.Biophys.Acta, 1838, 2014
|
|
8FS5
 
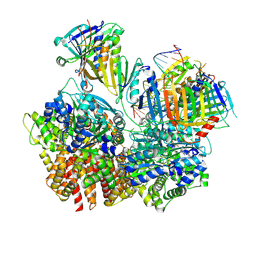 | | Structure of S. cerevisiae Rad24-RFC loading the 9-1-1 clamp onto a 10-nt gapped DNA in step 3 (open 9-1-1 and stably bound chamber DNA) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Checkpoint protein RAD24, DDC1 isoform 1, ... | | Authors: | Zheng, F, Georgescu, R, Yao, Y.N, O'Donnell, M.E, Li, H. | | Deposit date: | 2023-01-09 | | Release date: | 2023-06-14 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Structures of 9-1-1 DNA checkpoint clamp loading at gaps from start to finish and ramification to biology.
Biorxiv, 2023
|
|
8FS4
 
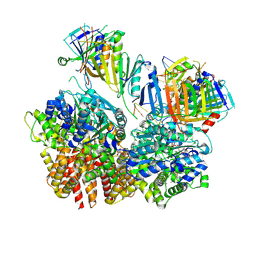 | | Structure of S. cerevisiae Rad24-RFC loading the 9-1-1 clamp onto a 10-nt gapped DNA in step 2 (open 9-1-1 ring and flexibly bound chamber DNA) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Checkpoint protein RAD24, DDC1 isoform 1, ... | | Authors: | Zheng, F, Georgescu, R, Yao, Y.N, O'Donnell, M.E, Li, H. | | Deposit date: | 2023-01-09 | | Release date: | 2023-06-14 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | Structures of 9-1-1 DNA checkpoint clamp loading at gaps from start to finish and ramification to biology.
Biorxiv, 2023
|
|
8FS3
 
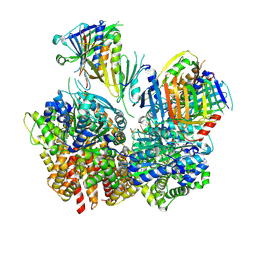 | | Structure of S. cerevisiae Rad24-RFC loading the 9-1-1 clamp onto a 10-nt gapped DNA in step 1 (open 9-1-1 and shoulder bound DNA only) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Checkpoint protein RAD24, DDC1 isoform 1, ... | | Authors: | Zheng, F, Georgescu, R, Yao, Y.N, O'Donnell, M.E, Li, H. | | Deposit date: | 2023-01-09 | | Release date: | 2023-06-14 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Structures of 9-1-1 DNA checkpoint clamp loading at gaps from start to finish and ramification to biology.
Biorxiv, 2023
|
|
8FS8
 
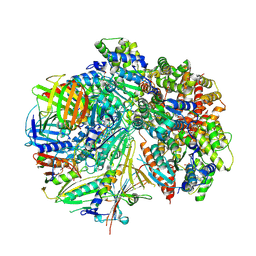 | | Structure of S. cerevisiae Rad24-RFC loading the 9-1-1 clamp onto a 5-nt gapped DNA (9-1-1 encircling fully bound DNA) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Checkpoint protein RAD24, DDC1 isoform 1, ... | | Authors: | Zheng, F, Georgescu, R, Yao, Y.N, O'Donnell, M.E, Li, H. | | Deposit date: | 2023-01-09 | | Release date: | 2023-06-14 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Structures of 9-1-1 DNA checkpoint clamp loading at gaps from start to finish and ramification to biology.
Biorxiv, 2023
|
|
8FS7
 
 | | Structure of S. cerevisiae Rad24-RFC loading the 9-1-1 clamp onto a 10-nt gapped DNA in step 5 (closed 9-1-1 and stably bound chamber DNA) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Checkpoint protein RAD24, DDC1 isoform 1, ... | | Authors: | Zheng, F, Georgescu, R, Yao, Y.N, O'Donnell, M.E, Li, H. | | Deposit date: | 2023-01-09 | | Release date: | 2023-06-14 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Structures of 9-1-1 DNA checkpoint clamp loading at gaps from start to finish and ramification to biology.
Biorxiv, 2023
|
|
8FS6
 
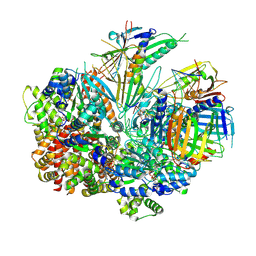 | | Structure of S. cerevisiae Rad24-RFC loading the 9-1-1 clamp onto a 10-nt gapped DNA in step 4 (partially closed 9-1-1 and stably bound chamber DNA) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Checkpoint protein RAD24, DDC1 isoform 1, ... | | Authors: | Zheng, F, Georgescu, R, Yao, Y.N, O'Donnell, M.E, Li, H. | | Deposit date: | 2023-01-09 | | Release date: | 2023-06-14 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structures of 9-1-1 DNA checkpoint clamp loading at gaps from start to finish and ramification to biology.
Biorxiv, 2023
|
|
6DEG
 
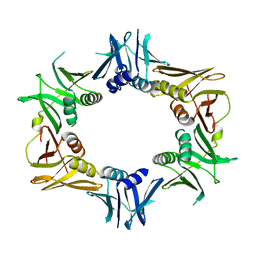 | |
4MXP
 
 | | Structural Basis for PI(4)P-Specific Membrane Recruitment of the Legionella pneumophila Effector DrrA/SidM | | Descriptor: | (2R)-3-{[(R)-hydroxy{[(1R,2R,3R,4R,5S,6R)-2,3,5,6-tetrahydroxy-4-(phosphonooxy)cyclohexyl]oxy}phosphoryl]oxy}propane-1,2-diyl dibutanoate, Defects in Rab1 recruitment protein A, SODIUM ION | | Authors: | Del Campo, C.M, Mishra, A.K, Wang, Y.H, Roy, C.R, Janmey, P.A, Lambright, D.G. | | Deposit date: | 2013-09-26 | | Release date: | 2014-03-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structural Basis for PI(4)P-Specific Membrane Recruitment of the Legionella pneumophila Effector DrrA/SidM.
Structure, 22, 2014
|
|
6VS0
 
 | | protein B | | Descriptor: | CHLORAMPHENICOL, Multidrug transporter MdfA, PRASEODYMIUM ION | | Authors: | Lu, M, Lu, M.M. | | Deposit date: | 2020-02-10 | | Release date: | 2020-06-03 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and mechanism of a redesigned multidrug transporter from the Major Facilitator Superfamily.
Sci Rep, 10, 2020
|
|
6VRZ
 
 | | protein A | | Descriptor: | CHLORAMPHENICOL, Multidrug transporter MdfA, PRASEODYMIUM ION | | Authors: | Lu, M, Lu, M.M. | | Deposit date: | 2020-02-10 | | Release date: | 2020-06-03 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and mechanism of a redesigned multidrug transporter from the Major Facilitator Superfamily.
Sci Rep, 10, 2020
|
|
4IRU
 
 | | Crystal Structure of lepB GAP core in a transition state mimetic complex with Rab1A and ALF3 | | Descriptor: | ACETATE ION, ALUMINUM FLUORIDE, GLYCEROL, ... | | Authors: | Mishra, A.K, Delcampo, C.M, Collins, R.E, Roy, C.R, Lambright, D.G. | | Deposit date: | 2013-01-15 | | Release date: | 2013-07-10 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The Legionella pneumophila GTPase Activating Protein LepB Accelerates Rab1 Deactivation by a Non-canonical Hydrolytic Mechanism.
J.Biol.Chem., 288, 2013
|
|
4CF2
 
 | | Interrogating HIV integrase for compounds that bind- a SAMPL challenge | | Descriptor: | (2s)-6-[[(1r,2s)-2-(4-azanylbutanoylamino)-2,3-dihydro-1h-inden-1-yl]methyl]-2-(3-hydroxy-3-oxopropyl)-2,3-dihydro-1,4-benzodioxine-5-carboxylic acid, INTEGRASE, SULFATE ION | | Authors: | Peat, T.S. | | Deposit date: | 2013-11-13 | | Release date: | 2013-11-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Interrogating HIV Integrase for Compounds that Bind- a Sampl Challenge.
J.Comput.Aided Mol.Des., 28, 2014
|
|
4CF9
 
 | | Interrogating HIV integrase for compounds that bind- a SAMPL challenge | | Descriptor: | (2R)-2-(3-hydroxy-3-oxopropyl)-6-[(E)-[(2S)-2-oxidanyl-2,3-dihydroinden-1-ylidene]methyl]-2,3-dihydro-1,4-benzodioxine-5-carboxylic acid, (3R)-3-(3-hydroxy-3-oxopropyl)-6-[(E)-[(2R)-2-oxidanyl-2,3-dihydroinden-1-ylidene]methyl]-2,3-dihydro-1,4-benzodioxine-5-carboxylic acid, 1,2-ETHANEDIOL, ... | | Authors: | Peat, T.S. | | Deposit date: | 2013-11-14 | | Release date: | 2013-11-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Interrogating HIV Integrase for Compounds that Bind- a Sampl Challenge.
J.Comput.Aided Mol.Des., 28, 2014
|
|
4CEZ
 
 | | Interrogating HIV integrase for compounds that bind- a SAMPL challenge | | Descriptor: | (2S)-2-(2-carboxyethyl)-6-{[{2-[(cyclohexylmethyl)carbamoyl]benzyl}(prop-2-en-1-yl)amino]methyl}-2,3-dihydro-1,4-benzodioxine-5-carboxylic acid, CHLORIDE ION, GLYCEROL, ... | | Authors: | Peat, T.S. | | Deposit date: | 2013-11-13 | | Release date: | 2013-11-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Interrogating HIV Integrase for Compounds that Bind- a Sampl Challenge.
J.Comput.Aided Mol.Des., 28, 2014
|
|
4CF1
 
 | | Interrogating HIV integrase for compounds that bind- a SAMPL challenge | | Descriptor: | (2s)-6-[[(1r,2s)-2-(5-azanylpentanoylamino)-2,3-dihydro-1h-inden-1-yl]methyl]-2-(3-hydroxy-3-oxopropyl)-2,3-dihydro-1,4-benzodioxine-5-carboxylic acid, ACETATE ION, INTEGRASE, ... | | Authors: | Peat, T.S. | | Deposit date: | 2013-11-13 | | Release date: | 2013-12-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Interrogating HIV Integrase for Compounds that Bind- a Sampl Challenge.
J.Comput.Aided Mol.Des., 28, 2014
|
|
4CFB
 
 | | Interrogating HIV integrase for compounds that bind- a SAMPL challenge | | Descriptor: | (R)-(4-CARBOXY-1,3-BENZODIOXOL-5-YL)METHYL-[[2-(CYCLOHEXYLMETHYLCARBAMOYL)PHENYL]METHYL]-METHYL-AZANIUM, ACETATE ION, INTEGRASE, ... | | Authors: | Peat, T.S. | | Deposit date: | 2013-11-14 | | Release date: | 2013-11-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Interrogating HIV Integrase for Compounds that Bind- a Sampl Challenge.
J.Comput.Aided Mol.Des., 28, 2014
|
|
4CF0
 
 | | Interrogating HIV integrase for compounds that bind- a SAMPL challenge | | Descriptor: | (2R)-6-[[(1R,2S)-2-(6-azanylhexanoylamino)-2,3-dihydro-1H-inden-1-yl]methyl]-2-(3-hydroxy-3-oxopropyl)-2,3-dihydro-1,4-benzodioxine-5-carboxylic acid, ACETATE ION, INTEGRASE, ... | | Authors: | Peat, T.S. | | Deposit date: | 2013-11-13 | | Release date: | 2013-11-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Interrogating HIV Integrase for Compounds that Bind- a Sampl Challenge.
J.Comput.Aided Mol.Des., 28, 2014
|
|
4CFC
 
 | | Interrogating HIV integrase for compounds that bind- a SAMPL challenge | | Descriptor: | 5-[[[(1S)-2-(butylamino)-2-oxidanylidene-1-phenyl-ethyl]carbamoyl-methyl-amino]methyl]-1,3-benzodioxole-4-carboxylic acid, ACETATE ION, CHLORIDE ION, ... | | Authors: | Peat, T.S. | | Deposit date: | 2013-11-14 | | Release date: | 2013-11-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Interrogating HIV Integrase for Compounds that Bind- a Sampl Challenge.
J.Comput.Aided Mol.Des., 28, 2014
|
|
6D57
 
 | | Campylobacter jejuni ferric uptake regulator S1 metalated | | Descriptor: | FORMIC ACID, Ferric uptake regulation protein, GLYCEROL, ... | | Authors: | Sarvan, S, Brunzelle, J.S, Couture, J.F. | | Deposit date: | 2018-04-19 | | Release date: | 2018-05-23 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Functional insights into the interplay between DNA interaction and metal coordination in ferric uptake regulators.
Sci Rep, 8, 2018
|
|
