6FJW
 
 | |
7UAQ
 
 | | Structure of the SARS-CoV-2 NTD in complex with C1520, local refinement | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, C1520 Fab Heavy Chain, ... | | Authors: | Barnes, C.O. | | Deposit date: | 2022-03-13 | | Release date: | 2022-04-27 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Analysis of memory B cells identifies conserved neutralizing epitopes on the N-terminal domain of variant SARS-Cov-2 spike proteins.
Immunity, 55, 2022
|
|
8DCB
 
 | | RNA ligase RtcB from Pyrococcus horikoshii in complex with Ni2+ and GTP | | Descriptor: | CHLORIDE ION, GLYCEROL, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Jacewicz, A, Dantuluri, S, Shuman, S. | | Deposit date: | 2022-06-16 | | Release date: | 2022-10-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of RNA ligase RtcB in complexes with divalent cations and GTP.
Rna, 28, 2022
|
|
6XM4
 
 | | Structure of SARS-CoV-2 spike at pH 5.5, single RBD up, conformation 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhou, T, Tsybovsky, Y, Olia, A, Kwong, P.D. | | Deposit date: | 2020-06-29 | | Release date: | 2020-08-12 | | Last modified: | 2021-12-15 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM Structures of SARS-CoV-2 Spike without and with ACE2 Reveal a pH-Dependent Switch to Mediate Endosomal Positioning of Receptor-Binding Domains.
Cell Host Microbe, 28, 2020
|
|
5HCC
 
 | | Ternary complex of human Complement C5 with Ornithodoros moubata OmCI and Dermacentor andersoni RaCI3. | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIETHYLENE DIOXIDE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Jore, M.M, Johnson, S, Lea, S.M. | | Deposit date: | 2016-01-04 | | Release date: | 2016-03-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Structural basis for therapeutic inhibition of complement C5.
Nat.Struct.Mol.Biol., 23, 2016
|
|
6FGG
 
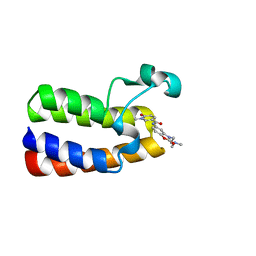 | | Crystal Structure of BAZ2A bromodomain in complex with 1-methylpyridinone compound 5 | | Descriptor: | Bromodomain adjacent to zinc finger domain protein 2A, ~{N}-[3-[2-(dimethylamino)ethyl]-2-oxidanylidene-1,3-benzoxazol-5-yl]-1-methyl-6-oxidanylidene-pyridine-3-carboxamide | | Authors: | Dalle Vedove, A, Spiliotopoulos, D, Lolli, G, Caflisch, A. | | Deposit date: | 2018-01-10 | | Release date: | 2018-05-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structural Analysis of Small-Molecule Binding to the BAZ2A and BAZ2B Bromodomains.
ChemMedChem, 13, 2018
|
|
8DCF
 
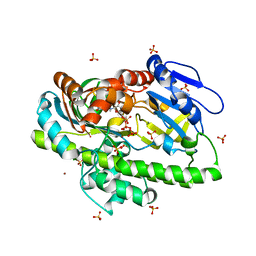 | | RNA ligase RtcB from Pyrococcus horikoshii in complex with Cu2+ and GTP | | Descriptor: | CHLORIDE ION, COPPER (II) ION, GLYCEROL, ... | | Authors: | Jacewicz, A, Dantuluri, S, Shuman, S. | | Deposit date: | 2022-06-16 | | Release date: | 2022-10-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structures of RNA ligase RtcB in complexes with divalent cations and GTP.
Rna, 28, 2022
|
|
6WQZ
 
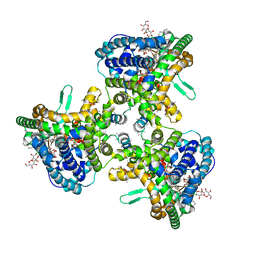 | | Structure of human ATG9A, the only transmembrane protein of the core autophagy machinery | | Descriptor: | Autophagy-related protein 9A, Lauryl Maltose Neopentyl Glycol | | Authors: | Guardia, C.M, Tan, X, Lian, T, Rana, M.S, Zhou, W, Christenson, E.T, Lowry, A.J, Faraldo-Gomez, J.D, Bonifacino, J.S, Jiang, J, Banerjee, A. | | Deposit date: | 2020-04-29 | | Release date: | 2020-07-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structure of Human ATG9A, the Only Transmembrane Protein of the Core Autophagy Machinery.
Cell Rep, 31, 2020
|
|
5K1V
 
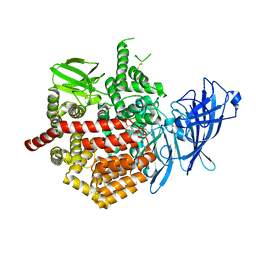 | | Crystal structure of Endoplasmic Reticulum aminopeptidase 2 (ERAP2) in complex with a diaminobenzoic acid derivative ligand. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Endoplasmic reticulum aminopeptidase 2, ... | | Authors: | Saridakis, E, Papakyriakou, A, Giastas, P, Mpakali, A, Mavridis, I.M, Stratikos, E. | | Deposit date: | 2016-05-18 | | Release date: | 2017-03-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.897 Å) | | Cite: | Crystal Structures of ERAP2 Complexed with Inhibitors Reveal Pharmacophore Requirements for Optimizing Inhibitor Potency.
ACS Med Chem Lett, 8, 2017
|
|
6A5E
 
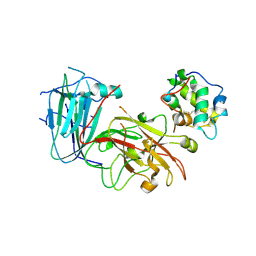 | | Crystal structure of plant peptide RALF23 in complex with FER and LLG2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GPI-anchored protein LLG2, ... | | Authors: | Xiao, Y, Chai, J. | | Deposit date: | 2018-06-23 | | Release date: | 2019-06-26 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.766 Å) | | Cite: | Mechanisms of RALF peptide perception by a heterotypic receptor complex.
Nature, 572, 2019
|
|
8DCA
 
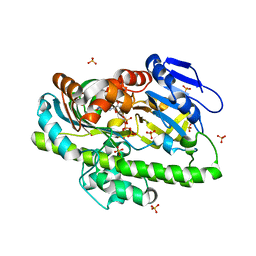 | | RNA ligase RtcB from Pyrococcus horikoshii in complex with Co2+ and GTP | | Descriptor: | COBALT (II) ION, GLYCEROL, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Jacewicz, A, Dantuluri, S, Shuman, S. | | Deposit date: | 2022-06-16 | | Release date: | 2022-10-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structures of RNA ligase RtcB in complexes with divalent cations and GTP.
Rna, 28, 2022
|
|
6ORP
 
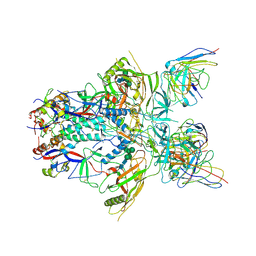 | | Modified BG505 SOSIP-based immunogen RC1 in complex with the elicited V3-glycan patch antibody Ab897NHP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Ab897NHP antibody Fab heavy chain, ... | | Authors: | Abernathy, M.E, Gristick, H.B, Bjorkman, P.J. | | Deposit date: | 2019-04-30 | | Release date: | 2019-06-12 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Immunization expands B cells specific to HIV-1 V3 glycan in mice and macaques.
Nature, 570, 2019
|
|
4JDT
 
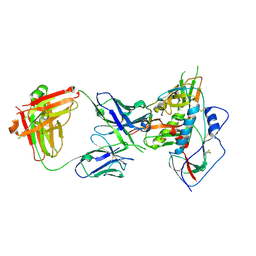 | | Crystal structure of chimeric germ-line precursor of NIH45-46 Fab in complex with gp120 of 93TH057 HIV-1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab heavy chain, Fab light chain, ... | | Authors: | Scharf, L, Diskin, R, Bjorkman, P.J. | | Deposit date: | 2013-02-25 | | Release date: | 2013-03-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.26 Å) | | Cite: | Structural basis for HIV-1 gp120 recognition by a germ-line version of a broadly neutralizing antibody.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
5K6W
 
 | | Sidekick-1 immunoglobulin domains 1-5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Goodman, K.M, Mannepalli, S, Honig, B, Shapiro, L. | | Deposit date: | 2016-05-25 | | Release date: | 2016-09-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Molecular basis of sidekick-mediated cell-cell adhesion and specificity.
Elife, 5, 2016
|
|
1E6N
 
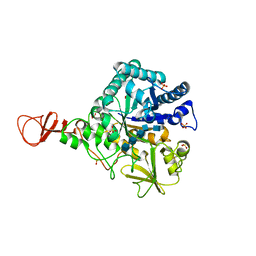 | | Chitinase B from Serratia marcescens inactive mutant E144Q in complex with N-acetylglucosamine-pentamer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHITINASE B, GLYCEROL, ... | | Authors: | Komander, D, Synstad, B, Eijsink, V.G.H, Van Aalten, D.M.F. | | Deposit date: | 2000-08-21 | | Release date: | 2001-06-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural Insights Into the Catalytic Mechanism of a Family 18 Exo-Chitinase
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
5JON
 
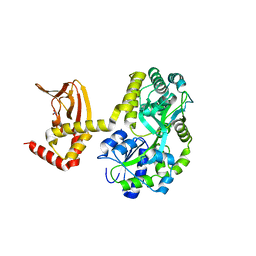 | | Crystal structure of the unliganded form of HCN2 CNBD | | Descriptor: | Maltose-binding periplasmic protein,Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2, NITRATE ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Klenchin, V.A, Chanda, B. | | Deposit date: | 2016-05-02 | | Release date: | 2016-11-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.042 Å) | | Cite: | Structure and dynamics underlying elementary ligand binding events in human pacemaking channels.
Elife, 5, 2016
|
|
8B97
 
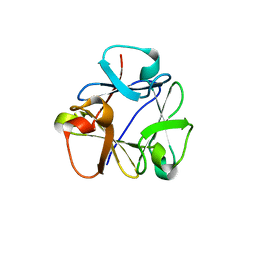 | | N-terminal beta-trefoil lectin domain of the Laccaria bicolor lectin in complex with N-acetyl-lactosamine | | Descriptor: | Beta-trefoil domain of the LBL lectin, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Acebron, I, Campanero-Rhodes, M.A, Solis, D, Menendez, M, Garcia, C, Lillo, M.P, Mancheno, J.M. | | Deposit date: | 2022-10-05 | | Release date: | 2023-02-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Atomic crystal structure and sugar specificity of a beta-trefoil lectin domain from the ectomycorrhizal basidiomycete Laccaria bicolor.
Int.J.Biol.Macromol., 233, 2023
|
|
7OR9
 
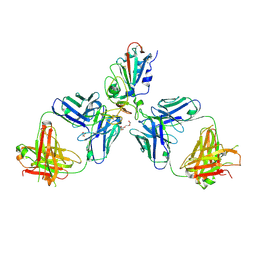 | | Crystal structure of the receptor binding domain of SARS-CoV-2 Spike glycoprotein in complex with COVOX-222 and COVOX-278 Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, COVOX-222 Fab heavy chain, ... | | Authors: | Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2021-06-04 | | Release date: | 2021-07-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Reduced neutralization of SARS-CoV-2 B.1.617 by vaccine and convalescent serum.
Cell, 184, 2021
|
|
7ORB
 
 | | Crystal structure of the L452R mutant receptor binding domain of SARS-CoV-2 Spike glycoprotein in complex with COVOX-75 and COVOX-253 Fabs | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2021-06-04 | | Release date: | 2021-07-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Reduced neutralization of SARS-CoV-2 B.1.617 by vaccine and convalescent serum.
Cell, 184, 2021
|
|
1ECY
 
 | | PROTEASE INHIBITOR ECOTIN | | Descriptor: | ECOTIN, alpha-D-glucopyranose, alpha-D-glucopyranose-(1-1)-alpha-D-glucopyranose, ... | | Authors: | Shin, D.H, Suh, S.W. | | Deposit date: | 1996-08-06 | | Release date: | 1997-02-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Crystal structure analyses of uncomplexed ecotin in two crystal forms: implications for its function and stability.
Protein Sci., 5, 1996
|
|
5JIK
 
 | | Crystal structure of HER2 binding IgG1-Fc (Fcab H10-03-6) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Ig gamma-1 chain C region, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Humm, A, Lobner, E, Mlynek, G, Obinger, C, Djinovic-Carugo, K. | | Deposit date: | 2016-04-22 | | Release date: | 2017-04-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Fcab-HER2 Interaction: a Menage a Trois. Lessons from X-Ray and Solution Studies.
Structure, 25, 2017
|
|
7ORA
 
 | | Crystal structure of the T478K mutant receptor binding domain of SARS-CoV-2 Spike glycoprotein in complex with COVOX-45 and COVOX-253 Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, COVOX-253 Fab heavy chain, ... | | Authors: | Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2021-06-04 | | Release date: | 2021-07-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Reduced neutralization of SARS-CoV-2 B.1.617 by vaccine and convalescent serum.
Cell, 184, 2021
|
|
6NB4
 
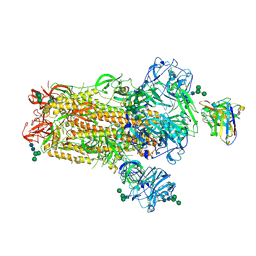 | | MERS-CoV S complex with human neutralizing LCA60 antibody Fab fragment (state 2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LCA60 heavy chain, ... | | Authors: | Walls, A.C, Xiong, X, Park, Y.J, Tortorici, M.A, Snijder, S, Quispe, J, Cameroni, E, Gopal, R, Mian, D, Lanzavecchia, A, Zambon, M, Rey, F.A, Corti, D, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2018-12-06 | | Release date: | 2019-02-06 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Unexpected Receptor Functional Mimicry Elucidates Activation of Coronavirus Fusion.
Cell, 176, 2019
|
|
7USB
 
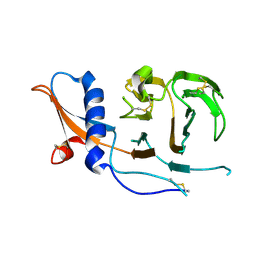 | | CCoV-HuPn-2018 S in the swung out conformation (local refinement of domain 0) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Tortorici, M.A, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2022-04-23 | | Release date: | 2022-08-24 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure, receptor recognition, and antigenicity of the human coronavirus CCoV-HuPn-2018 spike glycoprotein.
Cell, 185, 2022
|
|
7US9
 
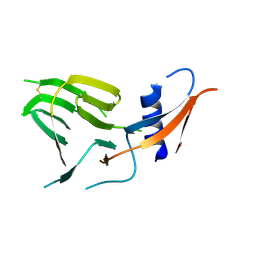 | | CCoV-HuPn-2018 S in the proximal conformation (local refinement of domain 0) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Tortorici, M.A, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2022-04-23 | | Release date: | 2022-08-24 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure, receptor recognition, and antigenicity of the human coronavirus CCoV-HuPn-2018 spike glycoprotein.
Cell, 185, 2022
|
|
