7WI4
 
 | | Cryo-EM structure of E.Coli FtsH protease cytosolic domains | | Descriptor: | ATP-dependent zinc metalloprotease FtsH, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Qiao, Z, Gao, Y.G. | | Deposit date: | 2022-01-02 | | Release date: | 2022-06-01 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structure of the entire FtsH-HflKC AAA protease complex.
Cell Rep, 39, 2022
|
|
1KIT
 
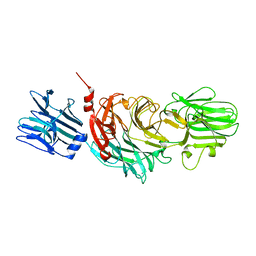 | | VIBRIO CHOLERAE NEURAMINIDASE | | Descriptor: | CALCIUM ION, SIALIDASE | | Authors: | Taylor, G.L, Crennell, S.J, Garman, E.F, Vimr, E.R, Laver, W.G. | | Deposit date: | 1996-06-21 | | Release date: | 1997-06-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of Vibrio cholerae neuraminidase reveals dual lectin-like domains in addition to the catalytic domain.
Structure, 2, 1994
|
|
2AZA
 
 | |
5CG9
 
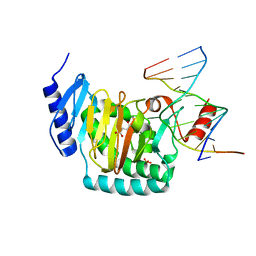 | | NgTET1 in complex with 5mC DNA in space group P3221 | | Descriptor: | 1,2-ETHANEDIOL, 2-OXOGLUTARIC ACID, DNA (5'-D(*TP*GP*TP*CP*AP*GP*(5CM)P*GP*CP*AP*TP*GP*G)-3'), ... | | Authors: | Hashimoto, H, Pais, J.E, Dai, N, Zhang, X, Zheng, Y, Cheng, X. | | Deposit date: | 2015-07-09 | | Release date: | 2015-09-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.693 Å) | | Cite: | Structure of Naegleria Tet-like dioxygenase (NgTet1) in complexes with a reaction intermediate 5-hydroxymethylcytosine DNA.
Nucleic Acids Res., 43, 2015
|
|
7BL4
 
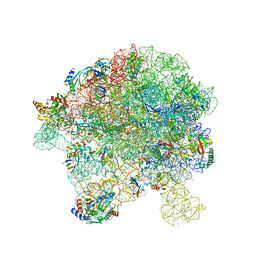 | | in vitro reconstituted 50S-ObgE-GMPPNP-RsfS particle | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Hilal, T, Nikolay, R, Spahn, C.M.T. | | Deposit date: | 2021-01-18 | | Release date: | 2021-05-12 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Snapshots of native pre-50S ribosomes reveal a biogenesis factor network and evolutionary specialization.
Mol.Cell, 81, 2021
|
|
4HCP
 
 | |
1E6R
 
 | | Chitinase B from Serratia marcescens wildtype in complex with inhibitor allosamidin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-allopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-allopyranose, ALLOSAMIZOLINE, CHITINASE B, ... | | Authors: | Komander, D, Synstad, B, Eijsink, V.G.H, Van Aalten, D.M.F. | | Deposit date: | 2000-08-22 | | Release date: | 2001-06-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Insights Into the Catalytic Mechanism of a Family 18 Exo-Chitinase
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
7BL2
 
 | | pre-50S-ObgE particle state 1 | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L11, 50S ribosomal protein L13, ... | | Authors: | Hilal, T, Nikolay, R, Schmidt, S, Spahn, C.M.T. | | Deposit date: | 2021-01-18 | | Release date: | 2021-05-12 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Snapshots of native pre-50S ribosomes reveal a biogenesis factor network and evolutionary specialization.
Mol.Cell, 81, 2021
|
|
2B1K
 
 | | Crystal structure of E. coli CcmG protein | | Descriptor: | Thiol:disulfide interchange protein dsbE | | Authors: | Ouyang, N, Gao, Y.G, Hu, H.Y, Xia, Z.X. | | Deposit date: | 2005-09-15 | | Release date: | 2006-09-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of E. coli CcmG and its mutants reveal key roles of the N-terminal beta-sheet and the fingerprint region
Proteins, 65, 2006
|
|
7BL6
 
 | | 50S-ObgE-GMPPNP particle | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Hilal, T, Nikolay, R, Schmidt, S, Spahn, C.M.T. | | Deposit date: | 2021-01-18 | | Release date: | 2021-05-12 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Snapshots of native pre-50S ribosomes reveal a biogenesis factor network and evolutionary specialization.
Mol.Cell, 81, 2021
|
|
1ZNH
 
 | | Strong Solute-Solute Dispersive Interactions in a Protein-Ligand Complex | | Descriptor: | CADMIUM ION, Major Urinary Protein, OCTAN-1-OL | | Authors: | Malham, R, Johnstone, S, Bingham, R.J, Barratt, E, Phillips, S.E, Laughton, C.A, Homans, S.W. | | Deposit date: | 2005-05-11 | | Release date: | 2005-12-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Strong Solute-Solute Dispersive Interactions in a Protein-Ligand Complex.
J.Am.Chem.Soc., 127, 2005
|
|
2AYA
 
 | |
7BL5
 
 | | pre-50S-ObgE particle | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L10, 50S ribosomal protein L11, ... | | Authors: | Hilal, T, Nikolay, R, Spahn, C.M.T, Schmidt, S. | | Deposit date: | 2021-01-18 | | Release date: | 2021-05-12 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Snapshots of native pre-50S ribosomes reveal a biogenesis factor network and evolutionary specialization.
Mol.Cell, 81, 2021
|
|
4HCS
 
 | | Structure of Novel subfamily CX chemokine solved by sulfur SAD | | Descriptor: | Uncharacterized protein | | Authors: | Rajasekaran, D, Fan, C, Meng, W, Pflugrath, J.W, Lolis, E.J. | | Deposit date: | 2012-10-01 | | Release date: | 2013-10-16 | | Last modified: | 2014-04-23 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Structural insight into the evolution of a new chemokine family from zebrafish.
Proteins, 82, 2014
|
|
7BL3
 
 | | pre-50S-ObgE particle state 2 | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L11, 50S ribosomal protein L13, ... | | Authors: | Hilal, T, Nikolay, R, Spahn, C.M.T. | | Deposit date: | 2021-01-18 | | Release date: | 2021-05-12 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Snapshots of native pre-50S ribosomes reveal a biogenesis factor network and evolutionary specialization.
Mol.Cell, 81, 2021
|
|
1ZOL
 
 | | native beta-PGM | | Descriptor: | MAGNESIUM ION, beta-phosphoglucomutase | | Authors: | Zhang, G, Tremblay, L.W, Dai, J, Wang, L, Dunaway-Mariano, D, Allen, K.N. | | Deposit date: | 2005-05-13 | | Release date: | 2005-08-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Catalytic cycling in beta-phosphoglucomutase: a kinetic and structural analysis
Biochemistry, 44, 2005
|
|
2B05
 
 | | Crystal Structure of 14-3-3 gamma in complex with a phosphoserine peptide | | Descriptor: | 14-3-3 protein gamma, peptide | | Authors: | Papagrigoriou, E, Elkins, J, Arrowsmith, C, Zhao, Y, Debreczeni, E.J, Edwards, A, Weigelt, J, Doyle, D, von Delft, F, Turnbull, A, Yang, X. | | Deposit date: | 2005-09-13 | | Release date: | 2005-10-11 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal Structure of 14-3-3 gamma in complex with a phosphoserine peptide
TO BE PUBLISHED
|
|
5CL4
 
 | |
1KHN
 
 | | E. COLI ALKALINE PHOSPHATASE MUTANT (D153HD330N) ZINC FORM | | Descriptor: | Alkaline phosphatase, ZINC ION | | Authors: | Le Du, M.H, Lamoure, C, Muller, B.H, Bulgakov, O.V, Lajeunesse, E, Menez, A, Boulain, J.C. | | Deposit date: | 2001-11-30 | | Release date: | 2002-03-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Artificial evolution of an enzyme active site: structural studies of three highly active mutants of Escherichia coli alkaline phosphatase.
J.Mol.Biol., 316, 2002
|
|
5CB5
 
 | | Structural Insights into the Mechanism of Escherichia coli Ymdb | | Descriptor: | ACETATE ION, ADENOSINE-5-DIPHOSPHORIBOSE, O-acetyl-ADP-ribose deacetylase, ... | | Authors: | Zhang, W, Wang, C, Song, Y, Shao, C, Zhang, X, Zang, J. | | Deposit date: | 2015-06-30 | | Release date: | 2015-11-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural insights into the mechanism of Escherichia coli YmdB: A 2'-O-acetyl-ADP-ribose deacetylase
J.Struct.Biol., 192, 2015
|
|
4KHA
 
 | | Structural basis of histone H2A-H2B recognition by the essential chaperone FACT | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, Histone H2A, ... | | Authors: | Hondele, M, Halbach, F, Hassler, M, Ladurner, A.G. | | Deposit date: | 2013-04-30 | | Release date: | 2013-05-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis of histone H2A-H2B recognition by the essential chaperone FACT.
Nature, 499, 2013
|
|
1E72
 
 | | Myrosinase from Sinapis alba with bound gluco-hydroximolactam and sulfate or ascorbate | | Descriptor: | (2S,3S,4R,5R)-6-(HYDROXYAMINO)-2-(HYDROXYMETHYL)-2,3,4,5-TETRAHYDROPYRIDINE-3,4,5-TRIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Burmeister, W.P. | | Deposit date: | 2000-08-23 | | Release date: | 2001-01-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | High resolution X-ray crystallography shows that ascorbate is a cofactor for myrosinase and substitutes for the function of the catalytic base.
J. Biol. Chem., 275, 2000
|
|
3ANP
 
 | | Crystal structure of Thermus thermophilus FadR, a TetR familly transcriptional repressor, in complex with lauroyl-CoA. | | Descriptor: | DODECYL-COA, LAURIC ACID, Transcriptional repressor, ... | | Authors: | Agari, Y, Agari, K, Sakamoto, K, Kuramitsu, S, Shinkai, A. | | Deposit date: | 2010-09-06 | | Release date: | 2011-03-09 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | TetR-family transcriptional repressor Thermus thermophilus FadR controls fatty acid degradation.
Microbiology, 157, 2011
|
|
3CSG
 
 | |
5CNB
 
 | | Ultrafast dynamics in myoglobin: 0.5 ps time delay | | Descriptor: | CARBON MONOXIDE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Barends, T.R.M, Foucar, L, Ardevol, A, Nass, K.J, Aquila, A, Botha, S, Doak, R.B, Falahati, K, Hartmann, E, Hilpert, M, Heinz, M, Hoffmann, M.C, Koefinger, J, Koglin, J, Kovacsova, G, Liang, M, Milathianaki, D, Lemke, H.T, Reinstein, J, Roome, C.M, Shoeman, R.L, Williams, G.J, Burghardt, I, Hummer, G, Boutet, S, Schlichting, I. | | Deposit date: | 2015-07-17 | | Release date: | 2015-09-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Direct observation of ultrafast collective motions in CO myoglobin upon ligand dissociation.
Science, 350, 2015
|
|
