4YJH
 
 | | Crystal structure of DAAO(Y228L/R283G) variant (R-2-phenylpyrrolidine binding form) | | Descriptor: | (2R)-2-phenylpyrrolidine, D-amino-acid oxidase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Nakano, S, Yasukawa, K, Kawahara, N, Ishitsubo, E, Tokiwa, H, Asano, Y. | | Deposit date: | 2015-03-03 | | Release date: | 2016-04-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of DAAO variant
To Be Published
|
|
1FPZ
 
 | | CRYSTAL STRUCTURE ANALYSIS OF KINASE ASSOCIATED PHOSPHATASE (KAP) WITH A SUBSTITUTION OF THE CATALYTIC SITE CYSTEINE (CYS140) TO A SERINE | | Descriptor: | CYCLIN-DEPENDENT KINASE INHIBITOR 3, SULFATE ION | | Authors: | Song, H, Hanlon, N, Brown, N.R, Noble, M.E.M, Johnson, L.N, Barford, D. | | Deposit date: | 2000-09-01 | | Release date: | 2001-05-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Phosphoprotein-protein interactions revealed by the crystal structure of kinase-associated phosphatase in complex with phosphoCDK2.
Mol.Cell, 7, 2001
|
|
4D4L
 
 | | human PFKFB3 in complex with a pyrrolopyrimidone compound | | Descriptor: | 5-(4-chlorophenyl)-7-phenyl-3,7-dihydro-4H-pyrrolo[2,3-d]pyrimidin-4-one, 6-O-phosphono-beta-D-fructofuranose, 6-PHOSPHOFRUCTO-2-KINASE/FRUCTOSE-2,6-BISPHOSPHATASE 3, ... | | Authors: | Stgallay, S.A, Bennett, N, Critchlow, S, Davies, G, Debreczeni, J.E, Evans, N, Holdgate, G, Jones, N.P, Leach, L, Maman, S, McLoughlin, S, Preston, M, Rigoreau, L, Thomas, A, Walker, G, Walsch, J, Ward, R.A, Wheatley, E, Winter, J. | | Deposit date: | 2014-10-29 | | Release date: | 2016-01-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.16 Å) | | Cite: | Identifying a Novel Series of Pfkfb3 Inhibitors as a Metabolic Approach to Treating Cancer from Hts, Biophysical and Biochemical Methods
To be Published
|
|
2V9I
 
 | |
6HKB
 
 | | Ternary complex of Estrogen Receptor alpha peptide and 14-3-3 sigma C42 mutant bound to disulfide fragment PPI stabilizer 3 | | Descriptor: | (1~{R},2~{S})-2-[methyl-[(~{R})-(2-methylpropan-2-yl)oxy-oxidanyl-methyl]amino]-2-phenyl-1-(2-sulfanylethylamino)ethanol, 14-3-3 protein sigma, Estrogen receptor, ... | | Authors: | Sijbesma, E, Hallenbeck, K.K, Leysen, S, Arkin, M.R, Ottmann, C. | | Deposit date: | 2018-09-06 | | Release date: | 2019-02-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Site-Directed Fragment-Based Screening for the Discovery of Protein-Protein Interaction Stabilizers.
J. Am. Chem. Soc., 141, 2019
|
|
1HJT
 
 | |
1BO4
 
 | | CRYSTAL STRUCTURE OF A GCN5-RELATED N-ACETYLTRANSFERASE: SERRATIA MARESCENS AMINOGLYCOSIDE 3-N-ACETYLTRANSFERASE | | Descriptor: | COENZYME A, PROTEIN (SERRATIA MARCESCENS AMINOGLYCOSIDE-3-N-ACETYLTRANSFERASE), SPERMIDINE | | Authors: | Wolf, E, Vassilev, A, Makino, Y, Sali, A, Nakatani, Y, Burley, S.K. | | Deposit date: | 1998-08-08 | | Release date: | 1998-10-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a GCN5-related N-acetyltransferase: Serratia marcescens aminoglycoside 3-N-acetyltransferase.
Cell(Cambridge,Mass.), 94, 1998
|
|
2NSP
 
 | | Crystal structure of pectin methylesterase D178A mutant in complex with hexasaccharide I | | Descriptor: | Pectinesterase A, methyl alpha-D-galactopyranuronate-(1-4)-methyl alpha-D-galactopyranuronate-(1-4)-alpha-D-galactopyranuronic acid-(1-4)-alpha-D-galactopyranuronic acid-(1-4)-alpha-D-galactopyranuronic acid-(1-4)-methyl alpha-D-galactopyranuronate | | Authors: | Fries, M, Brocklehurst, K, Shevchik, V.E, Pickersgill, R.W. | | Deposit date: | 2006-11-06 | | Release date: | 2007-09-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular basis of the activity of the phytopathogen pectin methylesterase.
Embo J., 26, 2007
|
|
1BL7
 
 | | THE COMPLEX STRUCTURE OF THE MAP KINASE P38/SB220025 | | Descriptor: | 4-(4-FLUOROPHENYL)-1-(4-PIPERIDINYL)-5-(2-AMINO-4-PYRIMIDINYL)-IMIDAZOLE, PROTEIN (MAP KINASE P38) | | Authors: | Wang, Z, Canagarajah, B.J, Boehm, J.C, Kassis, S, Cobb, M.H, Young, P.R, Abdel-Meguid, S, Adams, J.L, Goldsmith, E.J. | | Deposit date: | 1998-07-23 | | Release date: | 1999-07-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of inhibitor selectivity in MAP kinases.
Structure, 6, 1998
|
|
2IZS
 
 | | Structure of casein kinase gamma 3 in complex with inhibitor | | Descriptor: | CASEIN KINASE I ISOFORM GAMMA-3, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Bunkoczi, G, Salah, E, Rellos, P, Das, S, Fedorov, O, Savitsky, P, Debreczeni, J.E, Gileadi, O, Sundstrom, M, Edwards, A, Arrowsmith, C, Weigelt, J, von Delft, F, Knapp, S. | | Deposit date: | 2006-07-26 | | Release date: | 2006-08-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Inhibitor Binding by Casein Kinases
To be Published
|
|
2NT8
 
 | | ATP bound at the active site of a PduO type ATP:co(I)rrinoid adenosyltransferase from Lactobacillus reuteri | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cobalamin adenosyltransferase, GLYCEROL, ... | | Authors: | St-Maurice, M, Mera, P.E, Taranto, M.P, Sesma, F, Escalante-Semerena, J.C, Rayment, I. | | Deposit date: | 2006-11-07 | | Release date: | 2006-11-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural characterization of the active site of the PduO-type ATP:Co(I)rrinoid adenosyltransferase from Lactobacillus reuteri.
J.Biol.Chem., 282, 2007
|
|
3N0T
 
 | | Human dipeptidil peptidase DPP7 complexed with inhibitor GSK237826A | | Descriptor: | (3S)-4-oxo-4-piperidin-1-ylbutane-1,3-diamine, Dipeptidyl peptidase 2 | | Authors: | Dobrovetsky, E, Khutoreskaya, G, Seitova, A, Crombet, L, Cossar, D, Pagannon, S, Arrowsmith, C.H, Bountra, C, Weigelt, J, Edwards, A.M, Hassell, A, Shewchuk, L, Haffner, C, Bochkarev, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-05-14 | | Release date: | 2010-07-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Human dipeptidyl peptidase DPP7
To be Published
|
|
3N29
 
 | | Crystal structure of carboxynorspermidine decarboxylase complexed with Norspermidine from Campylobacter jejuni | | Descriptor: | Carboxynorspermidine decarboxylase, GLYCEROL, N-(3-aminopropyl)propane-1,3-diamine, ... | | Authors: | Deng, X, Lee, J, Michael, A.J, Tomchick, D.R, Goldsmith, E.J, Phillips, M.A. | | Deposit date: | 2010-05-17 | | Release date: | 2010-06-09 | | Last modified: | 2012-02-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Evolution of substrate specificity within a diverse family of beta/alpha-barrel-fold basic amino acid decarboxylases: X-ray structure determination of enzymes with specificity for L-arginine and carboxynorspermidine.
J.Biol.Chem., 285, 2010
|
|
2CA5
 
 | | MxiH needle protein of Shigella Flexneri (monomeric form, residues 1- 78) | | Descriptor: | GLYCEROL, ISOPROPYL ALCOHOL, MXIH, ... | | Authors: | Deane, J.E, Roversi, P, Cordes, F.S, Johnson, S, Kenjale, R, Picking, W.L, Picking, W.D, Blocker, A.J, Lea, S.M. | | Deposit date: | 2005-12-16 | | Release date: | 2006-08-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular Model of a Type III Secretion System Needle: Implications for Host-Cell Sensing
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
2NTT
 
 | | Crystal Structure of SEK | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Staphylococcal enterotoxin K | | Authors: | Gunther, S, Varma, A.K, Moza, B, Sundberg, E.J. | | Deposit date: | 2006-11-08 | | Release date: | 2007-06-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.561 Å) | | Cite: | A novel loop domain in superantigens extends their T cell receptor recognition site
J.Mol.Biol., 371, 2007
|
|
2C6R
 
 | | FE-SOAKED CRYSTAL STRUCTURE OF THE DPS92 FROM DEINOCOCCUS RADIODURANS | | Descriptor: | CHLORIDE ION, DNA-BINDING STRESS RESPONSE PROTEIN, DPS FAMILY, ... | | Authors: | Cuypers, M.G, Romao, C.V, Mitchell, E, Mcsweeney, S. | | Deposit date: | 2005-11-11 | | Release date: | 2007-02-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Crystal Structure of the Dps2 from Deinococcus Radiodurans Reveals an Unusual Pore Profile with a Non-Specific Metal Binding Site.
J.Mol.Biol., 371, 2007
|
|
2O84
 
 | | Crystal structure of K206E mutant of N-lobe human transferrin | | Descriptor: | CARBONATE ION, FE (III) ION, POTASSIUM ION, ... | | Authors: | Baker, H.M, Nurizzo, D, Mason, A.B, Baker, E.N. | | Deposit date: | 2006-12-12 | | Release date: | 2007-01-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of two mutants that probe the role in iron release of the dilysine pair in the N-lobe of human transferrin.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
4KWS
 
 | | Crystal structure of d-mannonate dehydratase from chromohalobacter salexigens complexed with Mg and glycerol | | Descriptor: | CHLORIDE ION, D-mannonate dehydratase, GLYCEROL, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Wichelecki, D, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2013-05-24 | | Release date: | 2014-05-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.642 Å) | | Cite: | Crystal structure of d-mannonate dehydratase from chromohalobacter salexigens complexed with Mg and glycerol
To be Published
|
|
2V9F
 
 | |
6A76
 
 | | Crystal structure of the Fab fragment of B5209B, a murine monoclonal antibody specific for the fifth immunoglobulin domain (Ig5) of human ROBO1 | | Descriptor: | GLYCEROL, Heavy chain of the anti-human Robo1 antibody B5209B Fab, Light chain of the anti-human Robo1 antibody B5209B Fab, ... | | Authors: | Mizohata, E, Nakayama, T, Kado, Y, Inoue, T. | | Deposit date: | 2018-07-02 | | Release date: | 2019-01-30 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Affinity Improvement of a Cancer-Targeted Antibody through Alanine-Induced Adjustment of Antigen-Antibody Interface.
Structure, 27, 2019
|
|
2VD9
 
 | | The crystal structure of alanine racemase from Bacillus anthracis (BA0252) with bound L-Ala-P | | Descriptor: | (1S)-1-[((1E)-{3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYLENE)AMINO]ETHYLPHOSPHONIC ACID, ALANINE RACEMASE, CHLORIDE ION, ... | | Authors: | Au, K, Ren, J, Walter, T.S, Harlos, K, Nettleship, J.E, Owens, R.J, Stuart, D.I, Esnouf, R.M, Oxford Protein Production Facility (OPPF), Structural Proteomics in Europe (SPINE) | | Deposit date: | 2007-10-01 | | Release date: | 2008-05-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of an Alanine Racemase from Bacillus Anthracis (Ba0252) in the Presence and Absence of (R)-1-Aminoethylphosphonic Acid (L-Ala-P).
Acta Crystallogr.,Sect.F, 64, 2008
|
|
1OUY
 
 | | The structure of p38 alpha in complex with a dihydropyrido-pyrimidine inhibitor | | Descriptor: | 1-(2,6-DICHLOROPHENYL)-6-[(2,4-DIFLUOROPHENYL)SULFANYL]-7-(1,2,3,6-TETRAHYDRO-4-PYRIDINYL)-3,4-DIHYDROPYRIDO[3,2-D]PYRIMIDIN-2(1H)-ONE, Mitogen-activated protein kinase 14 | | Authors: | Fitzgerald, C.E, Patel, S.B, Becker, J.W, Cameron, P.M, Zaller, D, Pikounis, V.B, O'Keefe, S.J, Scapin, G. | | Deposit date: | 2003-03-25 | | Release date: | 2003-09-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for p38alpha MAP kinase quinazolinone and pyridol-pyrimidine inhibitor specificity
Nat.Struct.Biol., 10, 2003
|
|
1RIH
 
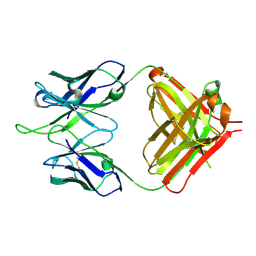 | | Crystal Structure of Fab 14F7, a unique anti-tumor antibody specific for N-glycolyl GM3 | | Descriptor: | heavy chain of antibody 14F7, light chain of antibody 14F7 | | Authors: | Krengel, U, Olsson, L.-L, Martinez, C, Talavera, A, Rojas, G, Mier, E, Angstrom, J, Moreno, E. | | Deposit date: | 2003-11-17 | | Release date: | 2004-01-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure and Molecular Interactions of a Unique Antitumor Antibody Specific for N-Glycolyl GM3.
J.Biol.Chem., 279, 2004
|
|
1W9M
 
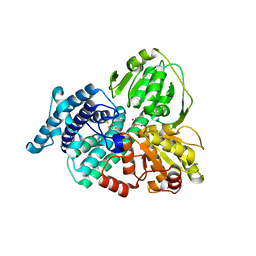 | | AS-isolated hybrid cluster protein from Desulfovibrio vulgaris X-ray structure at 1.35A resolution using iron anomalous signal | | Descriptor: | FE-S-O HYBRID CLUSTER, HYDROXYLAMINE REDUCTASE, IRON/SULFUR CLUSTER | | Authors: | Aragao, D, Macedo, S, Mitchell, E.P, Coelho, D, Romao, C.V, Teixeira, M, Lindley, P.F. | | Deposit date: | 2004-10-14 | | Release date: | 2005-02-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural and Functional Relationships in the Hybrid Cluster Protein Family:Structure of the Anaerobically Purified Hybrid Cluster Protein from Desulfovibrio Vulgaris at 1.35 A Resolution
Acta Crystallogr.,Sect.D, 64, 2008
|
|
2IJL
 
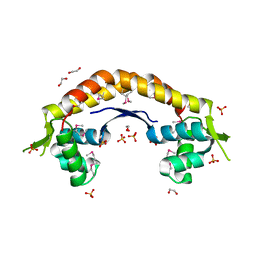 | | The structure of a putative ModE from Agrobacterium tumefaciens. | | Descriptor: | 1,2-ETHANEDIOL, Molybdenum-binding transcriptional repressor, SULFATE ION | | Authors: | Cuff, M.E, Evdokimova, E, Kudritska, M, Edwards, A, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-09-29 | | Release date: | 2006-10-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The structure of a putative ModE from Agrobacterium tumefaciens.
To be Published
|
|
