8Z9X
 
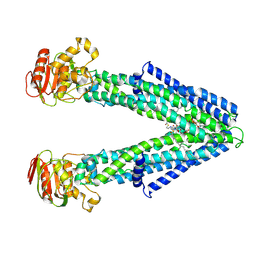 | |
9AX7
 
 | |
9CEZ
 
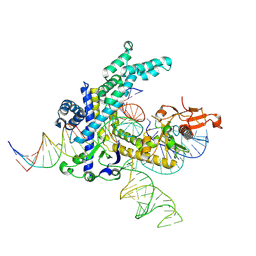 | | Spizellomyces punctatus Fanzor (SpuFz) State 6 | | Descriptor: | DNA (27-MER), DNA (5'-D(P*TP*AP*CP*CP*CP*GP*GP*GP*CP*AP*TP*A)-3'), MAGNESIUM ION, ... | | Authors: | Xu, P, Saito, M, Zhang, F. | | Deposit date: | 2024-06-27 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | Structural insights into the diversity and DNA cleavage mechanism of Fanzor.
Cell, 187, 2024
|
|
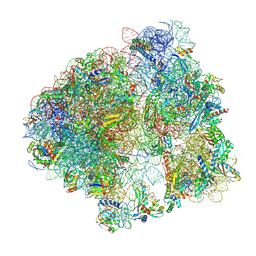
8ZHN
 
 | | SARS-CoV-2 spike trimer (6P) in complex with three H18 and three R1-32 Fabs (one RBD rotated) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of H18 Fab, ... | | Authors: | Yan, Q, Gao, X, Liu, B, Hou, R, He, P, Li, Z, Chen, Q, Wang, J, He, J, Chen, L, Zhao, J, Xiong, X. | | Deposit date: | 2024-05-11 | | Release date: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (4.12 Å) | | Cite: | Antibodies utilizing VL6-57 light chains target a convergent cryptic epitope on SARS-CoV-2 spike protein helping to drive the genesis of Omicron variants
To Be Published
|
|
9CG5
 
 | |
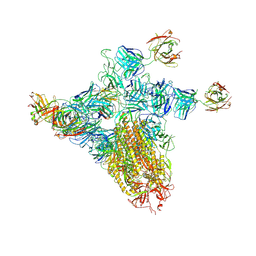
9CWS
 
 | | Bufavirus 1 at pH 2.6 | | Descriptor: | VP1 | | Authors: | Gulkis, M.C, McKenna, R, Bennett, A.D. | | Deposit date: | 2024-07-30 | | Release date: | 2024-08-21 | | Last modified: | 2024-09-18 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | Structural Characterization of Human Bufavirus 1: Receptor Binding and Endosomal pH-Induced Changes.
Viruses, 16, 2024
|
|
9FTI
 
 | | Open conformation of the pentameric ligand-gated ion channel, DeCLIC at pH 5 with 10mM EDTA | | Descriptor: | Neurotransmitter-gated ion-channel ligand-binding domain-containing protein | | Authors: | Rovsnik, U, Anden, O, Lycksell, M, Delarue, M, Howard, R.J, Lindahl, E. | | Deposit date: | 2024-06-24 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural characterization of pH-modulated closed and open states
in a pentameric ligand-gated ion channel
To Be Published
|
|
9EZY
 
 | | Vibrio cholerae DdmD-DdmE holo complex | | Descriptor: | 14 nucleotide DNA guide with terminal 5' phosphate, Helicase/UvrB N-terminal domain-containing protein, MAGNESIUM ION, ... | | Authors: | Loeff, L, Jinek, M. | | Deposit date: | 2024-04-14 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (2.56 Å) | | Cite: | Molecular mechanism of plasmid elimination by the DdmDE defense system.
Science, 385, 2024
|
|
9CG6
 
 | |
8Z4L
 
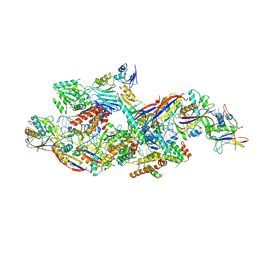 | | Cryo-EM structure of CTR-bound type VII CRISPR-Cas complex at substrate-engaged state I | | Descriptor: | RNA (40-MER), RNA (49-MER), ZINC ION, ... | | Authors: | Zhang, H, Deng, Z, Li, X. | | Deposit date: | 2024-04-17 | | Release date: | 2024-08-21 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Structural basis for the activity of the type VII CRISPR-Cas system.
Nature, 633, 2024
|
|
8Z9E
 
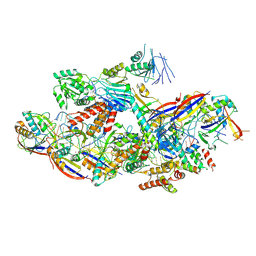 | | Cryo-EM structure of NTR-bound type VII CRISPR-Cas complex at substrate-engaged state II | | Descriptor: | Protein structure, RNA (34-MER), RNA (39-MER), ... | | Authors: | Zhang, H, Deng, Z, Li, X. | | Deposit date: | 2024-04-23 | | Release date: | 2024-08-21 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Structural basis for the activity of the type VII CRISPR-Cas system.
Nature, 633, 2024
|
|
9B1J
 
 | | Urate bound human URAT1 in the inward-facing state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PHOSPHATE ION, Solute carrier family 22 member 12, ... | | Authors: | Dai, Y, Lee, C.H. | | Deposit date: | 2024-03-13 | | Release date: | 2024-09-18 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Transport mechanism and structural pharmacology of human urate transporter URAT1.
Cell Res., 2024
|
|
8Z9C
 
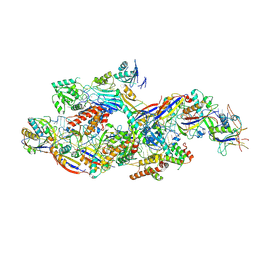 | | Cryo-EM structure of NTR-bound type VII CRISPR-Cas complex at substrate-engaged state I | | Descriptor: | Protein structure, RNA (41-MER), RNA (48-MER), ... | | Authors: | Zhang, H, Deng, Z, Li, X. | | Deposit date: | 2024-04-23 | | Release date: | 2024-08-21 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Structural basis for the activity of the type VII CRISPR-Cas system.
Nature, 633, 2024
|
|
9BDD
 
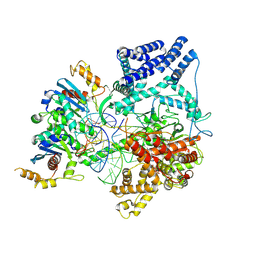 | | Cryo-EM Structure of Non-Cognate Substrate Bound in the Entry Site (ES) of Human Mitochondrial Transcription Elongation Complex | | Descriptor: | DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, DNA-directed RNA polymerase, mitochondrial, ... | | Authors: | Herbine, K.H, Nayak, A.R, Temiakov, D. | | Deposit date: | 2024-04-11 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Structural basis for substrate binding and selection by human mitochondrial RNA polymerase.
Nat Commun, 15, 2024
|
|
8ZYN
 
 | | Cryo-EM Structure of inhibitor-free hERG Channel | | Descriptor: | Potassium voltage-gated channel subfamily H member 2 | | Authors: | Miyashita, Y, Moriya, T, Kato, T, Kawasaki, M, Yasuda, Y, Adachi, N, Suzuki, K, Ogasawara, S, Saito, T, Senda, T, Murata, T. | | Deposit date: | 2024-06-18 | | Release date: | 2024-09-18 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Improved higher resolution cryo-EM structures reveal the binding modes of hERG channel inhibitors.
Structure, 2024
|
|
9B1L
 
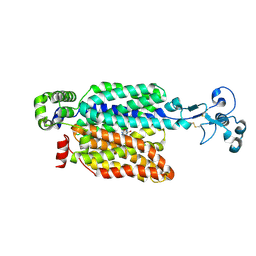 | | Urate bound human URAT1 in the outward-facing state. | | Descriptor: | Solute carrier family 22 member 12, URIC ACID | | Authors: | Dai, Y, Lee, C.H. | | Deposit date: | 2024-03-13 | | Release date: | 2024-09-18 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Transport mechanism and structural pharmacology of human urate transporter URAT1.
Cell Res., 2024
|
|
9CG7
 
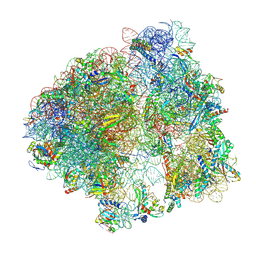 | |
9GGQ
 
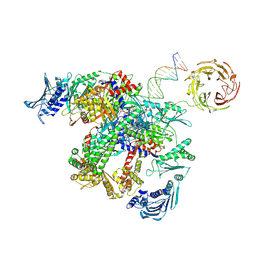 | | E.coli gyrase holocomplex with cleaved chirally wrapped 217 bp DNA fragment and moxifloxacin | | Descriptor: | 1-cyclopropyl-6-fluoro-8-methoxy-7-[(4aS,7aS)-octahydro-6H-pyrrolo[3,4-b]pyridin-6-yl]-4-oxo-1,4-dihydroquinoline-3-carboxylic acid, DNA gyrase subunit A, DNA gyrase subunit B, ... | | Authors: | Ghilarov, D, Heddle, J.G, Pabis, M. | | Deposit date: | 2024-08-13 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural basis of chiral wrap and T-segment capture by Escherichia coli DNA gyrase
Proceedings of the National Academy of Sciences USA, 2024
|
|
9BC4
 
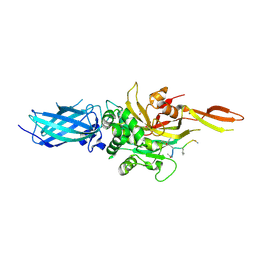 | | Transglutaminase 2 - Intermediate State | | Descriptor: | CALCIUM ION, GLYCEROL, HB-225 (gluten peptidomimetic TG2 inhibitor), ... | | Authors: | Sewa, A.S, Mathews, I.I, Khosla, C. | | Deposit date: | 2024-04-07 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structural and mechanistic analysis of Ca 2+ -dependent regulation of transglutaminase 2 activity using a Ca 2+ -bound intermediate state.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
9FOR
 
 | |
8Z5G
 
 | |
9F1C
 
 | | Mammalian quaternary complex of a translating 80S ribosome, NAC, MetAP1 and NatA/E | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S11, ... | | Authors: | Yudin, D, Scaiola, A, Ban, N. | | Deposit date: | 2024-04-18 | | Release date: | 2024-08-21 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3.78 Å) | | Cite: | NAC guides a ribosomal multienzyme complex for nascent protein processing.
Nature, 633, 2024
|
|
8ZYQ
 
 | | Cryo-EM Structure of pimozide-bound hERG Channel | | Descriptor: | 3-[1-[4,4-bis(4-fluorophenyl)butyl]piperidin-4-yl]-1~{H}-benzimidazol-2-one, Potassium voltage-gated channel subfamily H member 2 | | Authors: | Miyashita, Y, Moriya, T, Kato, T, Kawasaki, M, Yasuda, Y, Adachi, N, Suzuki, K, Ogasawara, S, Saito, T, Senda, T, Murata, T. | | Deposit date: | 2024-06-18 | | Release date: | 2024-09-18 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Improved higher resolution cryo-EM structures reveal the binding modes of hERG channel inhibitors.
Structure, 2024
|
|
8ZYC
 
 | |
9B9I
 
 | |
