7OMZ
 
 | |
7OMY
 
 | | Thermus sp. 2.9 DarT in complex with carba-NAD+ and ssDNA | | Descriptor: | CARBA-NICOTINAMIDE-ADENINE-DINUCLEOTIDE, DNA (5'-D(*AP*TP*GP*TP*C)-3'), DarT domain-containing protein, ... | | Authors: | Schuller, M, Ariza, A. | | Deposit date: | 2021-05-24 | | Release date: | 2021-06-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular basis for DarT ADP-ribosylation of a DNA base.
Nature, 596, 2021
|
|
8CMM
 
 | | Re-pairing DNA - binding of a ruthenium phi complex to a double mismatch | | Descriptor: | DNA (5'-D(*CP*GP*CP*TP*AP*TP*AP*AP*TP*GP*CP*G)-3'), LITHIUM ION, POTASSIUM ION, ... | | Authors: | Prieto Otoya, T.D, Cardin, C.J, McQuaid, K.M. | | Deposit date: | 2023-02-20 | | Release date: | 2024-03-06 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | Re-pairing DNA: binding of a ruthenium phi complex to a double mismatch.
Chem Sci, 15, 2024
|
|
2K4G
 
 | | Solution Structure of a Peptide Nucleic Acid Duplex, 10 structures | | Descriptor: | METHYLAMINE, PNA (N'-(*(GPN)*(GPN)*(CPN)*(APN)*(TPN)*(GPN)*(CPN)*(CPN))-C') | | Authors: | He, W, Hatcher, E, Balaeff, A, Beratan, D, Gil, R, Madrid, M, Achim, C. | | Deposit date: | 2008-06-07 | | Release date: | 2008-09-23 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a peptide nucleic acid duplex from NMR data: features and limitations.
J.Am.Chem.Soc., 130, 2008
|
|
8BNA
 
 | | BINDING OF HOECHST 33258 TO THE MINOR GROOVE OF B-DNA | | Descriptor: | 2'-(4-HYDROXYPHENYL)-5-(4-METHYL-1-PIPERAZINYL)-2,5'-BI-BENZIMIDAZOLE, DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3'), MAGNESIUM ION | | Authors: | Pjura, P.E, Grzeskowiak, K, Dickerson, R.E. | | Deposit date: | 1986-08-29 | | Release date: | 1987-01-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Binding of Hoechst 33258 to the minor groove of B-DNA.
J.Mol.Biol., 197, 1987
|
|
1JRN
 
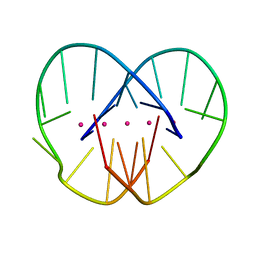 | |
1JPQ
 
 | |
6FQ2
 
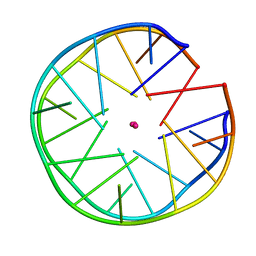 | | Structure of minimal sequence for left -handed G-quadruplex formation | | Descriptor: | DNA (5'-D(*GP*TP*GP*GP*TP*GP*GP*TP*GP*GP*TP*G)-3'), POTASSIUM ION | | Authors: | Schmitt, E, Mechulam, Y, Phan, A.T, Heddi, B, Bakalar, B. | | Deposit date: | 2018-02-13 | | Release date: | 2018-12-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | A Minimal Sequence for Left-Handed G-Quadruplex Formation.
Angew. Chem. Int. Ed. Engl., 58, 2019
|
|
1MWI
 
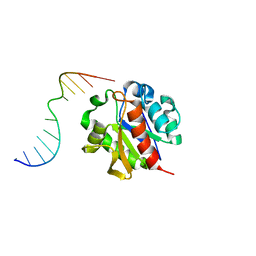 | | Crystal structure of a MUG-DNA product complex | | Descriptor: | 5'-D(*CP*GP*CP*GP*AP*GP*(AAB)P*TP*CP*GP*CP*G)-3', G/U mismatch-specific DNA glycosylase | | Authors: | Barrett, T.E, Savva, R, Panayotou, G, Brown, T, Barlow, T, Jiricny, J, Pearl, L.H. | | Deposit date: | 2002-09-30 | | Release date: | 2002-10-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of a G:T/U mismatch-specific DNA glycosylase: mismatch recognition by complementary-strand interactions.
Cell(Cambridge,Mass.), 92, 1998
|
|
6N88
 
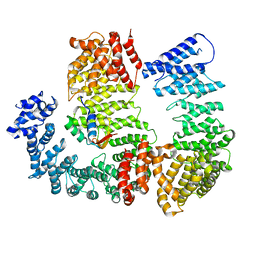 | |
3QOP
 
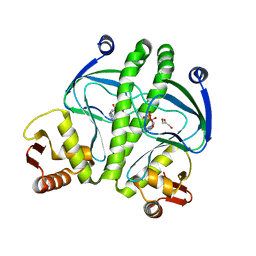 | |
1P7K
 
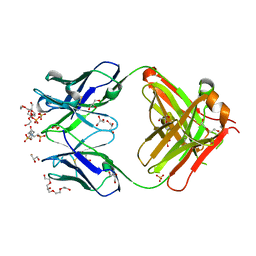 | | Crystal structure of an anti-ssDNA antigen-binding fragment (Fab) bound to 4-(2-Hydroxyethyl)piperazine-1-ethanesulfonic acid (HEPES) | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Schuermann, J.P, Henzl, M.T, Deutscher, S.L, Tanner, J.J. | | Deposit date: | 2003-05-02 | | Release date: | 2004-05-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of an anti-DNA fab complexed with a non-DNA ligand provides insights into cross-reactivity and molecular mimicry.
Proteins, 57, 2004
|
|
1KCI
 
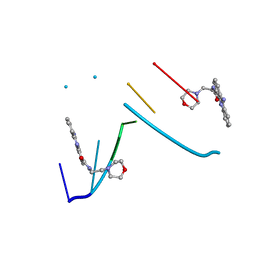 | | Crystal Structure of 9-amino-N-[2-(4-morpholinyl)ethyl]-4-acridinecarboxamide Bound to d(CGTACG)2 | | Descriptor: | 5'-D(*CP*GP*TP*AP*CP*G)-3', 9-AMINO-N-[2-(4-MORPHOLINYL)ETHYL]-4-ACRIDINECARBOXAMIDE | | Authors: | Adams, A, Guss, J.M, Denny, W.A, Wakelin, L.P.G. | | Deposit date: | 2001-11-08 | | Release date: | 2002-02-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of 9-amino-N-[2-(4-morpholinyl)ethyl]-4-acridinecarboxamide bound to d(CGTACG)2: implications for structure-activity relationships of acridinecarboxamide topoisomerase poisons.
Nucleic Acids Res., 30, 2002
|
|
3HIF
 
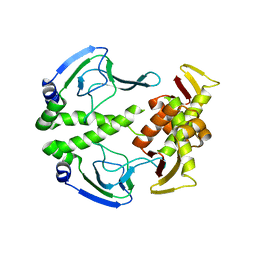 | | The crystal structure of apo wild type CAP at 3.6 A resolution. | | Descriptor: | Catabolite gene activator | | Authors: | Steitz, T.A, Sharma, H, Wang, J, Kong, J, Yu, S. | | Deposit date: | 2009-05-19 | | Release date: | 2009-09-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.59 Å) | | Cite: | Structure of apo-CAP reveals that large conformational changes are necessary for DNA binding.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3FWE
 
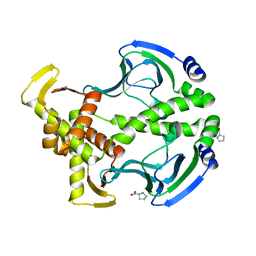 | | Crystal Structure of the Apo D138L CAP mutant | | Descriptor: | Catabolite gene activator, PROLINE | | Authors: | Sharma, H, Wang, J, Kong, J, Yu, S, Steitz, T. | | Deposit date: | 2009-01-17 | | Release date: | 2009-09-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of apo-CAP reveals that large conformational changes are necessary for DNA binding
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3ROU
 
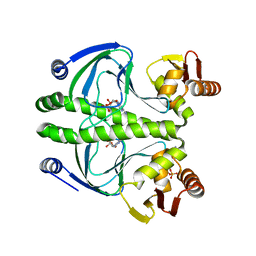 | |
3RPQ
 
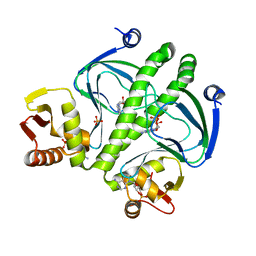 | |
3RDI
 
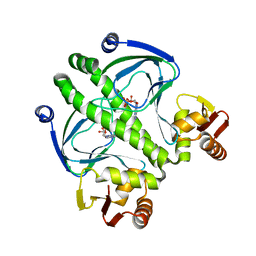 | |
3RYP
 
 | |
1D30
 
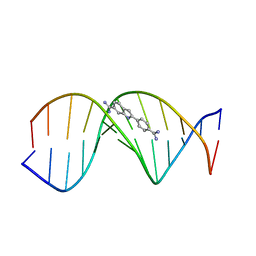 | | THE STRUCTURE OF DAPI BOUND TO DNA | | Descriptor: | 6-AMIDINE-2-(4-AMIDINO-PHENYL)INDOLE, DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3') | | Authors: | Larsen, T, Goodsell, D.S, Cascio, D, Grzeskowiak, K, Dickerson, R.E. | | Deposit date: | 1991-01-04 | | Release date: | 1992-04-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The structure of DAPI bound to DNA.
J.Biomol.Struct.Dyn., 7, 1989
|
|
6OD3
 
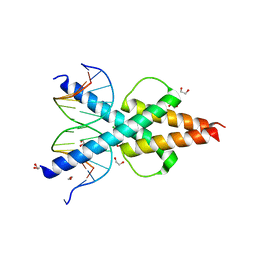 | | Human TCF4 C-terminal bHLH domain in Complex with 13-bp Oligonucleotide Containing E-box Sequence | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DNA (5'-D(*CP*AP*TP*AP*CP*AP*CP*GP*TP*GP*TP*AP*T)-3'), ... | | Authors: | Horton, J.R, Cheng, X, Yang, J. | | Deposit date: | 2019-03-25 | | Release date: | 2019-05-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.494 Å) | | Cite: | Structural basis for preferential binding of human TCF4 to DNA containing 5-carboxylcytosine.
Nucleic Acids Res., 47, 2019
|
|
1CR3
 
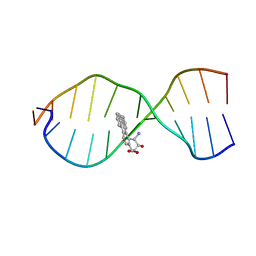 | | SOLUTION CONFORMATION OF THE (+)TRANS-ANTI-BENZO[G]CHRYSENE-DA ADDUCT OPPOSITE DT IN A DNA DUPLEX | | Descriptor: | BENZO[G]CHRYSENE, DNA (5'-D(*CP*TP*CP*TP*CP*AP*CP*TP*TP*CP*C)-3'), DNA (5'-D(*GP*GP*AP*AP*GP*TP*GP*AP*GP*AP*G)-3') | | Authors: | Suri, A.K, Mao, B, Amin, S, Geacintov, N.E, Patel, D.J. | | Deposit date: | 1999-08-12 | | Release date: | 2000-02-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution conformation of the (+)-trans-anti-benzo[g]chrysene-dA adduct opposite dT in a DNA duplex.
J.Mol.Biol., 292, 1999
|
|
1D44
 
 | | DNA DODECAMER C-G-C-G-A-A-T-T-C-G-C-G/HOECHST 33258 COMPLEX: 0 DEGREES C, PIPERAZINE DOWN | | Descriptor: | 2'-(4-HYDROXYPHENYL)-5-(4-METHYL-1-PIPERAZINYL)-2,5'-BI-BENZIMIDAZOLE, DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3'), MAGNESIUM ION | | Authors: | Quintana, J.R, Lipanov, A.A, Dickerson, R.E. | | Deposit date: | 1991-06-04 | | Release date: | 1992-04-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Low-temperature crystallographic analyses of the binding of Hoechst 33258 to the double-helical DNA dodecamer C-G-C-G-A-A-T-T-C-G-C-G.
Biochemistry, 30, 1991
|
|
3RYR
 
 | |
6OD4
 
 | | Human TCF4 C-terminal bHLH domain in Complex with 11-bp Oligonucleotide Containing E-box Sequence | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*TP*AP*CP*AP*CP*GP*TP*GP*TP*A)-3'), Transcription factor 4 | | Authors: | Horton, J.R, Cheng, X, Yang, J. | | Deposit date: | 2019-03-25 | | Release date: | 2019-05-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.699 Å) | | Cite: | Structural basis for preferential binding of human TCF4 to DNA containing 5-carboxylcytosine.
Nucleic Acids Res., 47, 2019
|
|
