5IDU
 
 | |
7APR
 
 | | Bacillithiol Disulfide Reductase Bdr (YpdA) from Staphylococcus aureus | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, YpdA family putative bacillithiol disulfide reductase Bdr | | Authors: | Hammerstad, M, Hersleth, H.-P. | | Deposit date: | 2020-10-19 | | Release date: | 2020-12-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The Crystal Structures of Bacillithiol Disulfide Reductase Bdr (YpdA) Provide Structural and Functional Insight into a New Type of FAD-Containing NADPH-Dependent Oxidoreductase.
Biochemistry, 59, 2020
|
|
5V36
 
 | | 1.88 Angstrom Resolution Crystal Structure of Glutathione Reductase from Streptococcus mutans UA159 in Complex with FAD | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Kiryukhina, O, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-03-06 | | Release date: | 2017-03-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | 1.88 Angstrom Resolution Crystal Structure of Glutathione Reductase from Streptococcus mutans UA159 in Complex with FAD.
To Be Published
|
|
3NTD
 
 | | Structure of the Shewanella loihica PV-4 NADH-dependent persulfide reductase C531S Mutant | | Descriptor: | CHLORIDE ION, COENZYME A, FAD-dependent pyridine nucleotide-disulphide oxidoreductase, ... | | Authors: | Sazinsky, M.H, Warner, M.D, Lukose, V, Lee, K.H, Crane, E.J. | | Deposit date: | 2010-07-03 | | Release date: | 2010-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Characterization of an NADH-Dependent Persulfide Reductase from Shewanella loihica PV-4: Implications for the Mechanism of Sulfur Respiration via FAD-Dependent Enzymes .
Biochemistry, 50, 2010
|
|
3NTA
 
 | | Structure of the Shewanella loihica PV-4 NADH-dependent persulfide reductase | | Descriptor: | CHLORIDE ION, COENZYME A, FAD-dependent pyridine nucleotide-disulphide oxidoreductase, ... | | Authors: | Sazinsky, M.H, Crane, E.J, Warner, M.D, Lukose, V, Lee, K.H. | | Deposit date: | 2010-07-03 | | Release date: | 2010-12-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Characterization of an NADH-Dependent Persulfide Reductase from Shewanella loihica PV-4: Implications for the Mechanism of Sulfur Respiration via FAD-Dependent Enzymes .
Biochemistry, 50, 2010
|
|
3NT6
 
 | | Structure of the Shewanella loihica PV-4 NADH-dependent persulfide reductase C43S/C531S Double Mutant | | Descriptor: | CHLORIDE ION, COENZYME A, FAD-dependent pyridine nucleotide-disulphide oxidoreductase, ... | | Authors: | Sazinsky, M.H, Crane, E.J, Warner, M.D, Lukose, V, Lee, K.H, Lopez, K. | | Deposit date: | 2010-07-02 | | Release date: | 2010-12-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Characterization of an NADH-Dependent Persulfide Reductase from Shewanella loihica PV-4: Implications for the Mechanism of Sulfur Respiration via FAD-Dependent Enzymes .
Biochemistry, 50, 2010
|
|
6NXJ
 
 | | Flavin Transferase ApbE from Vibrio cholerae, H257G mutant | | Descriptor: | FAD:protein FMN transferase, FLAVIN-ADENINE DINUCLEOTIDE, MAGNESIUM ION | | Authors: | Osipiuk, J, Fang, X, Chakravarthy, S, Juarez, O, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-02-08 | | Release date: | 2019-03-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Conserved residue His-257 ofVibrio choleraeflavin transferase ApbE plays a critical role in substrate binding and catalysis.
J.Biol.Chem., 294, 2019
|
|
4CY8
 
 | | 2-hydroxybiphenyl 3-monooxygenase (HbpA) in complex with FAD | | Descriptor: | 2-HYDROXYBIPHENYL 3-MONOOXYGENASE, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE | | Authors: | Jensen, C.N, Farrugia, J.E, Frank, A, Man, H, Hart, S, Turkenburg, J.P, Grogan, G. | | Deposit date: | 2014-04-10 | | Release date: | 2015-03-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structures of the Apo and Fad-Bound Forms of 2-Hydroxybiphenyl 3-Monooxygenase (Hbpa) Locate Activity Hotspots Identified by Using Directed Evolution.
Chembiochem, 16, 2015
|
|
4EH1
 
 | | Crystal Structure of the Flavohem-like-FAD/NAD Binding Domain of Nitric Oxide Dioxygenase from Vibrio cholerae O1 biovar El Tor | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, Flavohemoprotein, ... | | Authors: | Kim, Y, Gu, M, Hasseman, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-04-02 | | Release date: | 2012-04-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of the Flavohem-like-FAD/NAD Binding Domain of Nitric Oxide Dioxygenase from Vibrio cholerae O1 biovar El Tor
To be Published
|
|
7V0B
 
 | |
5TR9
 
 | |
4YNT
 
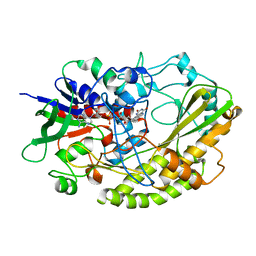 | | Crystal structure of Aspergillus flavus FAD glucose dehydrogenase | | Descriptor: | DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, Glucose oxidase, putative | | Authors: | Yoshida, H, Sakai, G, Kojima, K, Kamitori, S, Sode, K. | | Deposit date: | 2015-03-11 | | Release date: | 2015-09-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structural analysis of fungus-derived FAD glucose dehydrogenase
Sci Rep, 5, 2015
|
|
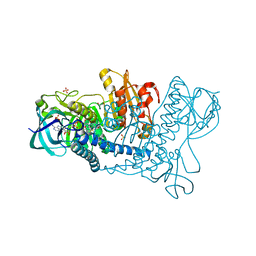
5U25
 
 | |
1KNR
 
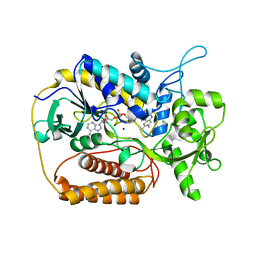 | | L-aspartate oxidase: R386L mutant | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, L-aspartate oxidase, ... | | Authors: | Bossi, R.T, Mattevi, A. | | Deposit date: | 2001-12-19 | | Release date: | 2002-04-17 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of FAD-bound L-aspartate oxidase: insight into substrate specificity and catalysis.
Biochemistry, 41, 2002
|
|
4NWZ
 
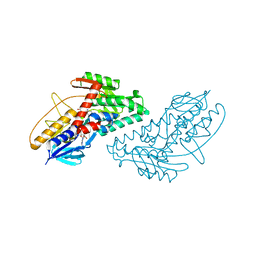 | | Structure of bacterial type II NADH dehydrogenase from Caldalkalibacillus thermarum at 2.5A resolution | | Descriptor: | FAD-dependent pyridine nucleotide-disulfide oxidoreductase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Nakatani, Y, Heikal, A, Lott, J.S, Sazanov, L.A, Baker, E.N, Cook, G.M. | | Deposit date: | 2013-12-07 | | Release date: | 2014-02-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the bacterial type II NADH dehydrogenase: a monotopic membrane protein with an essential role in energy generation.
Mol.Microbiol., 91, 2014
|
|
2RC6
 
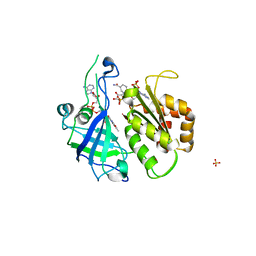 | | Refined structure of FNR from Leptospira interrogans bound to NADP+ | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Ferredoxin-NADP reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Nascimento, A.S, Catalano-Dupuy, D.L, Ceccarelli, E.A, Polikarpov, I. | | Deposit date: | 2007-09-19 | | Release date: | 2008-03-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of Leptospira interrogans FAD-containing ferredoxin-NADP+ reductase and its complex with NADP+.
Bmc Struct.Biol., 7, 2007
|
|
5FO0
 
 | |
5FO1
 
 | |
5FNZ
 
 | |
3OF4
 
 | |
1ZR6
 
 | | The crystal structure of an Acremonium strictum glucooligosaccharide oxidase reveals a novel flavinylation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FLAVIN-ADENINE DINUCLEOTIDE, ZINC ION, ... | | Authors: | Huang, C.-H, Lai, W.-L, Lee, M.-H, Tsai, Y.-C, Liaw, S.-H. | | Deposit date: | 2005-05-19 | | Release date: | 2005-09-13 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of glucooligosaccharide oxidase from Acremonium strictum: a novel flavinylation of 6-S-cysteinyl, 8alpha-N1-histidyl FAD
J.Biol.Chem., 280, 2005
|
|
3G5Q
 
 | | Crystal structure of Thermus thermophilus TrmFO | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, FLAVIN-ADENINE DINUCLEOTIDE, Methylenetetrahydrofolate--tRNA-(uracil-5-)-methyltransferase trmFO | | Authors: | Nishimasu, H, Ishitani, R, Hori, H, Nureki, O. | | Deposit date: | 2009-02-05 | | Release date: | 2009-05-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Atomic structure of a folate/FAD-dependent tRNA T54 methyltransferase
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3G5R
 
 | | Crystal structure of Thermus thermophilus TrmFO in complex with tetrahydrofolate | | Descriptor: | (6S)-5,6,7,8-TETRAHYDROFOLATE, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Nishimasu, H, Ishitani, R, Hori, H, Nureki, O. | | Deposit date: | 2009-02-05 | | Release date: | 2009-05-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Atomic structure of a folate/FAD-dependent tRNA T54 methyltransferase
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3G5S
 
 | | Crystal structure of Thermus thermophilus TrmFO in complex with glutathione | | Descriptor: | 1,2-ETHANEDIOL, FLAVIN-ADENINE DINUCLEOTIDE, GLUTATHIONE, ... | | Authors: | Nishimasu, H, Ishitani, R, Hori, H, Nureki, O. | | Deposit date: | 2009-02-05 | | Release date: | 2009-05-19 | | Last modified: | 2011-12-14 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Atomic structure of a folate/FAD-dependent tRNA T54 methyltransferase
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3U2M
 
 | | Crystal structure of human ALR mutant C142/145S | | Descriptor: | FAD-linked sulfhydryl oxidase ALR, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Banci, L, Bertini, I, Calderone, V, Cefaro, C, Ciofi-Baffoni, S, Gallo, A. | | Deposit date: | 2011-10-04 | | Release date: | 2012-06-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | An electron-transfer path through an extended disulfide relay system: the case of the redox protein ALR.
J.Am.Chem.Soc., 134, 2012
|
|
