4Z0G
 
 | | Structure of the IMPDH from Ashbya gossypii bound to GMP | | Descriptor: | GLYCEROL, GUANOSINE-5'-MONOPHOSPHATE, Inosine-5'-monophosphate dehydrogenase,Inosine-5'-monophosphate dehydrogenase, ... | | Authors: | Buey, R.M, de Pereda, J.M, Revuelta, J.L. | | Deposit date: | 2015-03-26 | | Release date: | 2015-11-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Guanine nucleotide binding to the Bateman domain mediates the allosteric inhibition of eukaryotic IMP dehydrogenases.
Nat Commun, 6, 2015
|
|
4XTI
 
 | | Structure of IMP dehydrogenase of Ashbya gossypii with IMP bound to the active site | | Descriptor: | INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase,Inosine-5'-monophosphate dehydrogenase, POTASSIUM ION | | Authors: | Buey, R.M, Ledesma-Amaro, R, Balsera, M, de Pereda, J.M, Revuelta, J.L. | | Deposit date: | 2015-01-23 | | Release date: | 2015-07-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Increased riboflavin production by manipulation of inosine 5'-monophosphate dehydrogenase in Ashbya gossypii.
Appl.Microbiol.Biotechnol., 99, 2015
|
|
4XWU
 
 | | Structure of the IMP dehydrogenase from Ashbya gossypii | | Descriptor: | Inosine-5'-monophosphate dehydrogenase,Inosine-5'-monophosphate dehydrogenase | | Authors: | Buey, R.M, Ledesma-Amaro, R, Balsera, M, de Pereda, J.M, Revuelta, J.L. | | Deposit date: | 2015-01-29 | | Release date: | 2015-07-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Increased riboflavin production by manipulation of inosine 5'-monophosphate dehydrogenase in Ashbya gossypii.
Appl.Microbiol.Biotechnol., 99, 2015
|
|
4XTD
 
 | | Structure of the covalent intermediate E-XMP* of the IMP dehydrogenase of Ashbya gossypii | | Descriptor: | INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase,Inosine-5'-monophosphate dehydrogenase | | Authors: | Buey, R.M, Ledesma-Amaro, R, Balsera, M, de Pereda, J.M, Revuelta, J.L. | | Deposit date: | 2015-01-23 | | Release date: | 2015-07-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Increased riboflavin production by manipulation of inosine 5'-monophosphate dehydrogenase in Ashbya gossypii.
Appl.Microbiol.Biotechnol., 99, 2015
|
|
6RPU
 
 | | Structure of the ternary complex of the IMPDH enzyme from Ashbya gossypii bound to the dinucleoside polyphosphate Ap5G and GDP | | Descriptor: | ACETATE ION, GUANOSINE-5'-DIPHOSPHATE, Inosine-5'-monophosphate dehydrogenase, ... | | Authors: | Buey, R.M, Fernandez-Justel, D, Revuelta, J.L. | | Deposit date: | 2019-05-14 | | Release date: | 2019-08-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | The Bateman domain of IMP dehydrogenase is a binding target for dinucleoside polyphosphates.
J.Biol.Chem., 294, 2019
|
|
4Z87
 
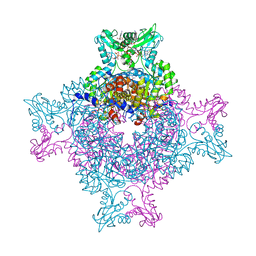 | | Structure of the IMP dehydrogenase from Ashbya gossypii bound to GDP | | Descriptor: | ACETATE ION, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-MONOPHOSPHATE, ... | | Authors: | Buey, R.M, de Pereda, J.M, Revuelta, J.L. | | Deposit date: | 2015-04-08 | | Release date: | 2015-11-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Guanine nucleotide binding to the Bateman domain mediates the allosteric inhibition of eukaryotic IMP dehydrogenases.
Nat Commun, 6, 2015
|
|
5MCP
 
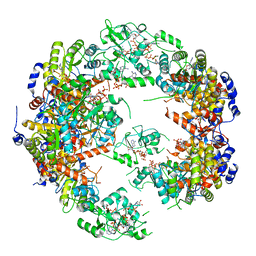 | | Structure of IMP dehydrogenase from Ashbya gossypii bound to ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Inosine-5'-monophosphate dehydrogenase, MAGNESIUM ION | | Authors: | Winter, G, Fernandez-Justel, D, de Pereda, J.M, Revuelta, J.L, Buey, R.M. | | Deposit date: | 2016-11-10 | | Release date: | 2017-06-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A nucleotide-controlled conformational switch modulates the activity of eukaryotic IMP dehydrogenases.
Sci Rep, 7, 2017
|
|
5TC3
 
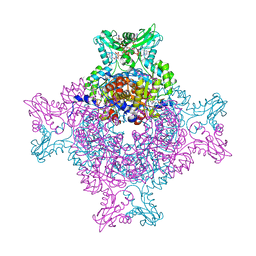 | | Structure of IMP dehydrogenase from Ashbya gossypii bound to ATP and GDP | | Descriptor: | ACETATE ION, ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Fernandez-Justel, D, de Pereda, J.M, Revuelta, J.L, Buey, R.M. | | Deposit date: | 2016-09-14 | | Release date: | 2017-06-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.462 Å) | | Cite: | A nucleotide-controlled conformational switch modulates the activity of eukaryotic IMP dehydrogenases.
Sci Rep, 7, 2017
|
|
