4MON
 
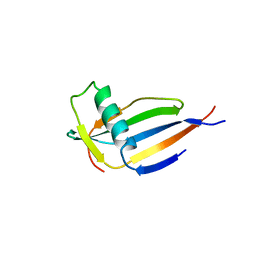 | | ORTHORHOMBIC MONELLIN | | Descriptor: | MONELLIN | | Authors: | Bujacz, G, Wlodawer, A. | | Deposit date: | 1997-03-04 | | Release date: | 1997-07-07 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of monellin refined to 2.3 a resolution in the orthorhombic crystal form.
Acta Crystallogr.,Sect.D, 53, 1997
|
|
3MON
 
 | |
7VWW
 
 | |
5XFU
 
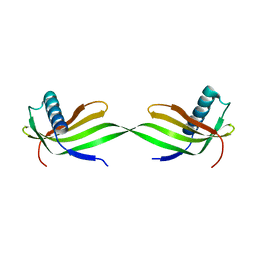 | | Domain swapped dimer crystal structure of loop1 deletion mutant in Single-chain Monellin | | Descriptor: | Monellin chain B,Monellin chain A | | Authors: | Surana, P, Nandwani, N, Udgaonkar, J, Gosavi, S, Das, R. | | Deposit date: | 2017-04-11 | | Release date: | 2017-07-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.611 Å) | | Cite: | Amino-acid composition after loop deletion drives domain swapping
Protein Sci., 26, 2017
|
|
2O9U
 
 | |
8TM8
 
 | |
8Q0R
 
 | | X-ray structure of MNEI mutant Mut9 (E23A, C41A, Y65R, S76Y) | | Descriptor: | ACETATE ION, Monellin chain B,Monellin chain A, SULFATE ION | | Authors: | Ferraro, G, Merlino, A, Lucignano, R, Picone, D. | | Deposit date: | 2023-07-29 | | Release date: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural insights and aggregation propensity of a super-stable monellin mutant: A new potential building block for protein-based nanostructured materials.
Int.J.Biol.Macromol., 254, 2024
|
|
8Q0S
 
 | | X-ray structure of the single chain monellin derivative MNEI | | Descriptor: | ACETATE ION, GLYCEROL, Monellin chain B,Monellin chain A, ... | | Authors: | Ferraro, G, Merlino, A, Lucignano, R, Picone, D. | | Deposit date: | 2023-07-29 | | Release date: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Structural insights and aggregation propensity of a super-stable monellin mutant: A new potential building block for protein-based nanostructured materials.
Int.J.Biol.Macromol., 254, 2024
|
|
8JZ0
 
 | |
8JZ1
 
 | |
5LC7
 
 | |
5LC6
 
 | |
7EUA
 
 | | X-ray structure of a P93A Monellin mutant | | Descriptor: | Monellin, SULFATE ION | | Authors: | Manjula, R, Bhatia, S, Jayant, B, Ramaswamy, S, Gosavi, S. | | Deposit date: | 2021-05-16 | | Release date: | 2021-08-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | X-ray structure of a P93A Monellin mutant
To Be Published
|
|
7D75
 
 | |
3PXM
 
 | | Reduced sweetness of a monellin (MNEI) mutant results from increased protein flexibility and disruption of a distant poly-(L-proline) II helix | | Descriptor: | Monellin chain B/Monellin chain A chimeric protein | | Authors: | Hobbs, J.R, Templeton, C.M, Pour, S.O, Blanch, E.W, Munger, S.M, Conn, G.L. | | Deposit date: | 2010-12-10 | | Release date: | 2011-04-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Reduced Sweetness of a Monellin (MNEI) Mutant Results from Increased Protein Flexibility and Disruption of a Distant Poly-(L-Proline) II Helix.
Chem Senses, 36, 2011
|
|
3PYJ
 
 | | Reduced sweetness of a monellin (MNEI) mutant results from increased protein flexibility and disruption of a distant poly-(L-proline) II helix | | Descriptor: | FORMIC ACID, Monellin chain B/Monellin chain A chimeric protein | | Authors: | Hobbs, J.R, Conn, G.C, Munger, S.D, Templeton, C.M. | | Deposit date: | 2010-12-13 | | Release date: | 2011-04-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Reduced Sweetness of a Monellin (MNEI) Mutant Results from Increased Protein Flexibility and Disruption of a Distant Poly-(L-Proline) II Helix.
Chem Senses, 36, 2011
|
|
3Q2P
 
 | | Reduced sweetness of a monellin (MNEI) mutant results from increased protein flexibility and disruption of a distant poly-(L-proline) II helix | | Descriptor: | Monellin chain B/Monellin chain A chimeric protein, SODIUM ION | | Authors: | Templeton, C.M, Hobbs, J.R, Munger, S.D, Conn, G.L. | | Deposit date: | 2010-12-20 | | Release date: | 2011-04-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.341 Å) | | Cite: | Reduced Sweetness of a Monellin (MNEI) Mutant Results from Increased Protein Flexibility and Disruption of a Distant Poly-(L-Proline) II Helix.
Chem Senses, 36, 2011
|
|
9HIK
 
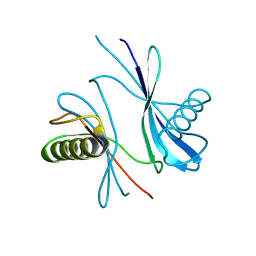 | | X-ray structure of Perm3, a circularly permuted mutant of the sweet protein MNEI | | Descriptor: | Monellin chain A,Monellin chain B | | Authors: | Bologna, A, Wang, P.-H, Lucignano, R, Essen, L.-O, Spadaccini, R. | | Deposit date: | 2024-11-26 | | Release date: | 2025-04-02 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Unravelling the amyloid aggregation mechanism of the sweet protein Monellin: Insights from circular permutated mutants.
Int.J.Biol.Macromol., 308, 2025
|
|
9HKG
 
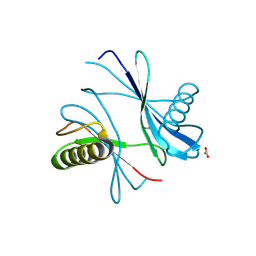 | | X-ray structure of Perm2, a circularly permuted mutant of the sweet protein MNEI | | Descriptor: | ACETIC ACID, Monellin chain A,Monellin chain B | | Authors: | Bologna, A, Wang, P.-H, Essen, L.-O, Spadaccini, R. | | Deposit date: | 2024-12-03 | | Release date: | 2025-04-02 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Unravelling the amyloid aggregation mechanism of the sweet protein Monellin: Insights from circular permutated mutants.
Int.J.Biol.Macromol., 308, 2025
|
|
9HIJ
 
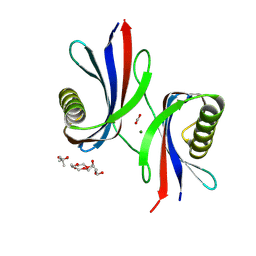 | | X-ray structure of Perm1, a circularly permuted mutant of the sweet protein MNEI | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, HEXAETHYLENE GLYCOL, ... | | Authors: | Bologna, A, Wang, P.-H, Gramazio, S, Essen, L.-O, Spadaccini, R. | | Deposit date: | 2024-11-26 | | Release date: | 2025-04-02 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Unravelling the amyloid aggregation mechanism of the sweet protein Monellin: Insights from circular permutated mutants.
Int.J.Biol.Macromol., 308, 2025
|
|
5YCU
 
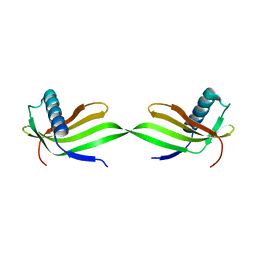 | | Domain swapped dimer of engineered hairpin loop1 mutant in Single-chain Monellin | | Descriptor: | Single chain monellin | | Authors: | Surana, P, Nandwani, N, Udgaonkar, J.B, Gosavi, S, Das, R. | | Deposit date: | 2017-09-08 | | Release date: | 2018-11-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | A five-residue motif for the design of domain swapping in proteins.
Nat Commun, 10, 2019
|
|
5YCW
 
 | | Double domain swapped dimer of engineered hairpin loop1 and loop3 mutant in Single-chain Monellin | | Descriptor: | single chain monellin | | Authors: | Surana, P, Nandwani, N, Udgaonkar, J.B, Gosavi, S, Das, R. | | Deposit date: | 2017-09-08 | | Release date: | 2018-11-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.285 Å) | | Cite: | A five-residue motif for the design of domain swapping in proteins.
Nat Commun, 10, 2019
|
|
1MNL
 
 | | HIGH-RESOLUTION SOLUTION STRUCTURE OF A SWEET PROTEIN SINGLE-CHAIN MONELLIN (SCM) DETERMINED BY NUCLEAR MAGNETIC RESONANCE SPECTROSCOPY AND DYNAMICAL SIMULATED ANNEALING CALCULATIONS, 21 STRUCTURES | | Descriptor: | MONELLIN | | Authors: | Lee, S.-Y, Lee, J.-H, Chang, H.-J, Jo, J.-M, Jung, J.-W, Lee, W. | | Deposit date: | 1998-08-06 | | Release date: | 1999-06-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a sweet protein single-chain monellin determined by nuclear magnetic resonance and dynamical simulated annealing calculations.
Biochemistry, 38, 1999
|
|
5YCT
 
 | | Engineered hairpin loop3 mutant monomer in Single-chain Monellin | | Descriptor: | Single chain Monellin | | Authors: | Surana, P, Nandwani, N, Udgaonkar, J.B, Gosavi, S, Das, R. | | Deposit date: | 2017-09-08 | | Release date: | 2018-11-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.851 Å) | | Cite: | A five-residue motif for the design of domain swapping in proteins.
Nat Commun, 10, 2019
|
|
1MOL
 
 | |
