1M12
 
 | |
5U85
 
 | | Murine saposin-D (SapD), open conformation | | Descriptor: | Saposin-D | | Authors: | Gebai, A, Gorelik, A, Illes, K, Nagar, B. | | Deposit date: | 2016-12-13 | | Release date: | 2018-06-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of saposin D in an open conformation.
J. Struct. Biol., 204, 2018
|
|
7P4T
 
 | | Tetrameric structure of murine SapA | | Descriptor: | Saposin-A | | Authors: | Shamin, M, Deane, J.E. | | Deposit date: | 2021-07-13 | | Release date: | 2022-05-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.17 Å) | | Cite: | A Tetrameric Assembly of Saposin A: Increasing Structural Diversity in Lipid Transfer Proteins.
Contact, 4, 2021
|
|
8EQU
 
 | | Structure of SARS-CoV-2 Orf3a in late endosome/lysosome-like environment, Saposin A nanodisc | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, ORF3a protein, Saposin A, ... | | Authors: | Miller, A.N, Houlihan, P.R, Matamala, E, Cabezas-Bratesco, D, Lee, G.Y, Cristofori-Armstrong, B, Dilan, T.L, Sanchez-Martinez, S, Matthies, D, Yan, R, Yu, Z, Ren, D, Brauchi, S.E, Clapham, D.E. | | Deposit date: | 2022-10-09 | | Release date: | 2023-02-08 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | The SARS-CoV-2 accessory protein Orf3a is not an ion channel, but does interact with trafficking proteins.
Elife, 12, 2023
|
|
5NXB
 
 | | Mouse galactocerebrosidase in complex with saposin A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Graham, S.C, Hill, C.H, Deane, J.E. | | Deposit date: | 2017-05-09 | | Release date: | 2017-05-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | The mechanism of glycosphingolipid degradation revealed by a GALC-SapA complex structure.
Nat Commun, 9, 2018
|
|
4UEX
 
 | |
2DOB
 
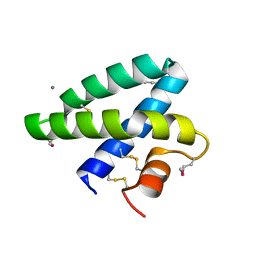 | | Crystal Structure of Human Saposin A | | Descriptor: | CALCIUM ION, Proactivator polypeptide | | Authors: | Prive, G.G, Ahn, V.E. | | Deposit date: | 2006-04-28 | | Release date: | 2006-07-25 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of saposins A and C.
Protein Sci., 15, 2006
|
|
9I63
 
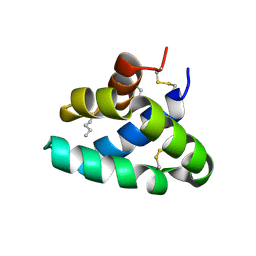 | | Synthetic Human Saposin D glycoprotein | | Descriptor: | Prosaposin, SULFATE ION | | Authors: | Weyand, M, Unverzagt, C. | | Deposit date: | 2025-01-29 | | Release date: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Synthetic Glycoforms Reveal Carbohydrate-Dependent Bioactivity of Human Saposin D.
Angew.Chem.Int.Ed.Engl., 56, 2017
|
|
4DDJ
 
 | |
7MBK
 
 | |
3RFI
 
 | |
2GTG
 
 | |
1SN6
 
 | |
2R1Q
 
 | |
1NKL
 
 | |
2RB3
 
 | | Crystal Structure of Human Saposin D | | Descriptor: | GLYCEROL, Proactivator polypeptide, SULFATE ION | | Authors: | Maier, T, Rossman, M, Saenger, W. | | Deposit date: | 2007-09-18 | | Release date: | 2008-04-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of human saposins C and d: implications for lipid recognition and membrane interactions.
Structure, 16, 2008
|
|
1QDM
 
 | | CRYSTAL STRUCTURE OF PROPHYTEPSIN, A ZYMOGEN OF A BARLEY VACUOLAR ASPARTIC PROTEINASE. | | Descriptor: | PROPHYTEPSIN | | Authors: | Kervinen, J, Tobin, G.J, Costa, J, Waugh, D.S, Wlodawer, A, Zdanov, A. | | Deposit date: | 1999-05-19 | | Release date: | 1999-07-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of plant aspartic proteinase prophytepsin: inactivation and vacuolar targeting.
EMBO J., 18, 1999
|
|
2QYP
 
 | |
2R0R
 
 | |
6VZD
 
 | | N-terminal domain of mouse surfactant protein B (K46E/R51E mutant) with bound lipid | | Descriptor: | (7Z,19R,22R)-25-amino-22-hydroxy-16,22-dioxo-17,21,23-trioxa-22lambda~5~-phosphapentacos-7-en-19-yl (9Z)-octadec-9-enoate, Pulmonary surfactant-associated protein B | | Authors: | Rapoport, T.A, Bodnar, N.O. | | Deposit date: | 2020-02-28 | | Release date: | 2020-11-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Mechanism of Lamellar Body Formation by Lung Surfactant Protein B.
Mol.Cell, 81, 2021
|
|
6VYN
 
 | | N-terminal domain of mouse surfactant protein B with bound lipid, wild type | | Descriptor: | (7Z,19R,22R)-25-amino-22-hydroxy-16,22-dioxo-17,21,23-trioxa-22lambda~5~-phosphapentacos-7-en-19-yl (9Z)-octadec-9-enoate, Pulmonary surfactant-associated protein B | | Authors: | Rapoport, T.A, Bodnar, N.O. | | Deposit date: | 2020-02-27 | | Release date: | 2020-11-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mechanism of Lamellar Body Formation by Lung Surfactant Protein B.
Mol.Cell, 81, 2021
|
|
6W1B
 
 | |
2Z9A
 
 | |
3BQP
 
 | |
3BQQ
 
 | |
