1Y8P
 
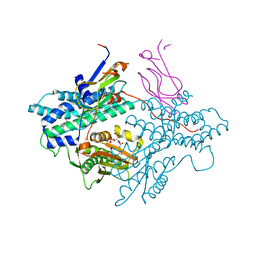 | | Crystal structure of the PDK3-L2 complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DIHYDROLIPOIC ACID, Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex, ... | | Authors: | Kato, M, Chuang, J.L, Wynn, R.M, Chuang, D.T. | | Deposit date: | 2004-12-13 | | Release date: | 2005-05-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Crystal structure of pyruvate dehydrogenase kinase 3 bound to lipoyl domain 2 of human pyruvate dehydrogenase complex.
Embo J., 24, 2005
|
|
1Y8N
 
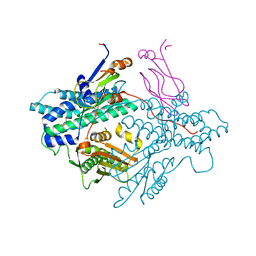 | | Crystal structure of the PDK3-L2 complex | | Descriptor: | DIHYDROLIPOIC ACID, Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex, POTASSIUM ION, ... | | Authors: | Kato, M, Chuang, J.L, Wynn, R.M, Chuang, D.T. | | Deposit date: | 2004-12-13 | | Release date: | 2005-05-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of pyruvate dehydrogenase kinase 3 bound to lipoyl domain 2 of human pyruvate dehydrogenase complex.
Embo J., 24, 2005
|
|
1Y8O
 
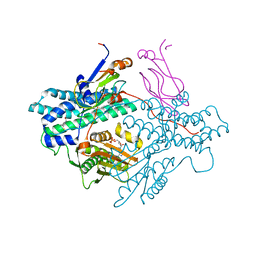 | | Crystal structure of the PDK3-L2 complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DIHYDROLIPOIC ACID, Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex, ... | | Authors: | Kato, M, Chuang, J.L, Wynn, R.M, Chuang, D.T. | | Deposit date: | 2004-12-13 | | Release date: | 2005-05-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Crystal structure of pyruvate dehydrogenase kinase 3 bound to lipoyl domain 2 of human pyruvate dehydrogenase complex.
Embo J., 24, 2005
|
|
1Y4U
 
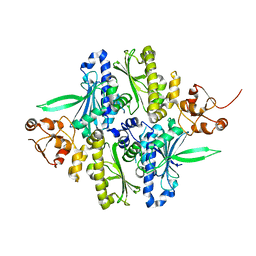 | | Conformation rearrangement of heat shock protein 90 upon ADP binding | | Descriptor: | Chaperone protein htpG | | Authors: | Huai, Q, Wang, H, Liu, Y, Kim, H, Toft, D, Ke, H. | | Deposit date: | 2004-12-01 | | Release date: | 2005-04-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of the N-terminal and middle domains of E. coli Hsp90 and conformation changes upon ADP binding.
Structure, 13, 2005
|
|
1Y4S
 
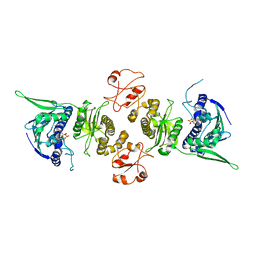 | | Conformation rearrangement of heat shock protein 90 upon ADP binding | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Chaperone protein htpG, MAGNESIUM ION | | Authors: | Huai, Q, Wang, H, Liu, Y, Kim, H, Toft, D, Ke, H. | | Deposit date: | 2004-12-01 | | Release date: | 2005-04-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of the N-terminal and middle domains of E. coli Hsp90 and conformation changes upon ADP binding.
Structure, 13, 2005
|
|
6ENG
 
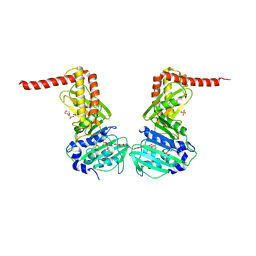 | | Crystal structure of the 43K ATPase domain of Escherichia coli gyrase B in complex with an aminocoumarin | | Descriptor: | CHLORIDE ION, Coumermycin A1, DNA gyrase subunit B, ... | | Authors: | Vanden Broeck, A, McEwen, A.G, Lamour, V. | | Deposit date: | 2017-10-04 | | Release date: | 2019-04-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis for DNA Gyrase Interaction with Coumermycin A1.
J.Med.Chem., 62, 2019
|
|
8QDX
 
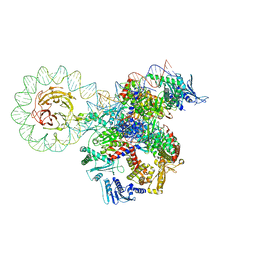 | | E. coli DNA gyrase bound to a DNA crossover | | Descriptor: | DNA gyrase subunit A, DNA gyrase subunit B, DNA minicircle | | Authors: | Vayssieres, M, Lamour, V. | | Deposit date: | 2023-08-30 | | Release date: | 2024-04-10 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of DNA crossover capture by Escherichia coli DNA gyrase.
Science, 384, 2024
|
|
8QQS
 
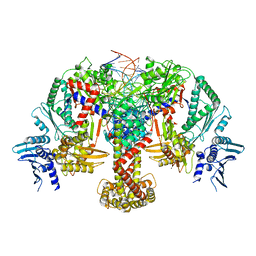 | |
8QQI
 
 | |
8SGK
 
 | | CryoEM structure of Deinococcus radiodurans BphP photosensory module in Pr state | | Descriptor: | 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium -2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3- yl]propanoic acid, Bacteriophytochrome | | Authors: | Li, H, Li, H. | | Deposit date: | 2023-04-12 | | Release date: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | CryoEM structure of Deinococcus radiodurans BphP photosensory module in Pr state
To Be Published
|
|
6GAU
 
 | | Extremely 'open' clamp structure of DNA gyrase: role of the Corynebacteriales GyrB specific insert | | Descriptor: | DNA gyrase subunit B,DNA gyrase subunit A, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Petrella, S, Capton, E, Alzari, P.M, Aubry, A, Mayer, C. | | Deposit date: | 2018-04-12 | | Release date: | 2019-02-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Overall Structures of Mycobacterium tuberculosis DNA Gyrase Reveal the Role of a Corynebacteriales GyrB-Specific Insert in ATPase Activity.
Structure, 27, 2019
|
|
8QQU
 
 | |
6GAV
 
 | | Extremely 'open' clamp structure of DNA gyrase: role of the Corynebacteriales GyrB specific insert | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DNA gyrase subunit B,DNA gyrase subunit A | | Authors: | Petrella, S, Capton, E, Alzari, P.M, Aubry, A, MAyer, C. | | Deposit date: | 2018-04-12 | | Release date: | 2019-02-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Overall Structures of Mycobacterium tuberculosis DNA Gyrase Reveal the Role of a Corynebacteriales GyrB-Specific Insert in ATPase Activity.
Structure, 27, 2019
|
|
2IOQ
 
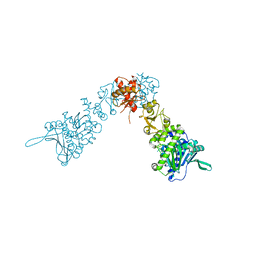 | |
2IOP
 
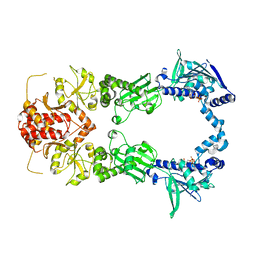 | | Crystal Structure of Full-length HtpG, the Escherichia coli Hsp90, Bound to ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Chaperone protein htpG | | Authors: | Shiau, A.K, Harris, S.F, Agard, D.A. | | Deposit date: | 2006-10-10 | | Release date: | 2006-11-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.55 Å) | | Cite: | Structural Analysis of E. coli hsp90 reveals dramatic nucleotide-dependent conformational rearrangements.
Cell(Cambridge,Mass.), 127, 2006
|
|
2PNR
 
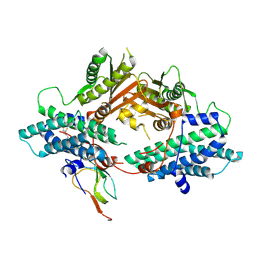 | | Crystal Structure of the asymmetric Pdk3-l2 Complex | | Descriptor: | DIHYDROLIPOIC ACID, Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex, [Pyruvate dehydrogenase [lipoamide]] kinase isozyme 3 | | Authors: | Vassylyev, D.G, Steussy, C.N, Devedjiev, Y. | | Deposit date: | 2007-04-25 | | Release date: | 2007-08-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of an Asymmetric complex of Pyruvate Dehydrogenase
Kinase 3 with Lipoyl domain 2 and its Biological Implications
J.Mol.Biol., 370, 2007
|
|
5J5Q
 
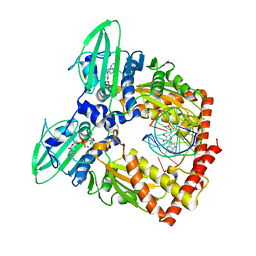 | | AMP-PNP-stabilized ATPase domain of topoisomerase IV from Streptococcus pneumoniae, complex type II | | Descriptor: | DNA (5'-D(*GP*CP*AP*TP*AP*TP*AP*TP*AP*TP*AP*TP*GP*C)-3'), DNA topoisomerase 4 subunit B, MAGNESIUM ION, ... | | Authors: | Laponogov, I, Pan, X.-S, Skamrova, G, Umrekar, T, Fisher, L.M, Sanderson, M.R. | | Deposit date: | 2016-04-03 | | Release date: | 2017-07-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Trapping of the transport-segment DNA by the ATPase domains of a type II topoisomerase.
Nat Commun, 9, 2018
|
|
8F71
 
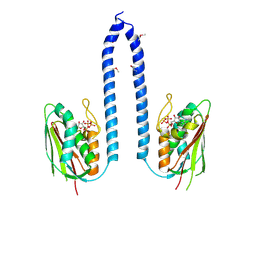 | |
5IUK
 
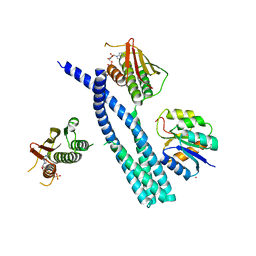 | | Crystal structure of the DesK-DesR complex in the phosphotransfer state with high Mg2+ (150 mM) | | Descriptor: | MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, POTASSIUM ION, ... | | Authors: | Trajtenberg, F, Imelio, J.A, Larrieux, N, Buschiazzo, A. | | Deposit date: | 2016-03-18 | | Release date: | 2016-12-21 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Regulation of signaling directionality revealed by 3D snapshots of a kinase:regulator complex in action.
Elife, 5, 2016
|
|
6LGQ
 
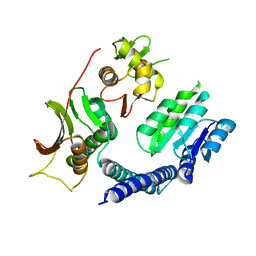 | |
8G3F
 
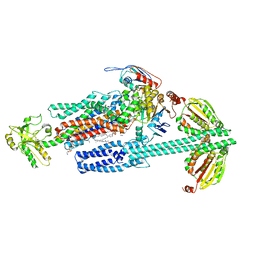 | | BceAB-S nucleotide free BceS state 1 | | Descriptor: | Bacitracin export ATP-binding protein BceA, Bacitracin export permease protein BceB, OLEIC ACID, ... | | Authors: | George, N.L, Orlando, B.J. | | Deposit date: | 2023-02-07 | | Release date: | 2023-06-21 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Architecture of a complete Bce-type antimicrobial peptide resistance module.
Nat Commun, 14, 2023
|
|
8G3B
 
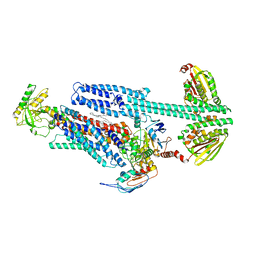 | | BceAB-S nucleotide free TM state 2 | | Descriptor: | Bacitracin export ATP-binding protein BceA, Bacitracin export permease protein BceB, OLEIC ACID, ... | | Authors: | George, N.L, Orlando, B.J. | | Deposit date: | 2023-02-07 | | Release date: | 2023-06-21 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Architecture of a complete Bce-type antimicrobial peptide resistance module.
Nat Commun, 14, 2023
|
|
8G3L
 
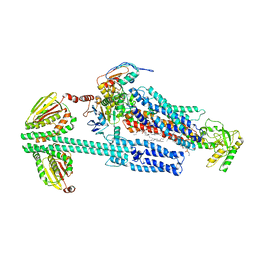 | | BceAB-S nucleotide free BceS state 2 | | Descriptor: | Bacitracin export ATP-binding protein BceA, Bacitracin export permease protein BceB, OLEIC ACID, ... | | Authors: | George, N.L, Orlando, B.J. | | Deposit date: | 2023-02-08 | | Release date: | 2023-06-21 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Architecture of a complete Bce-type antimicrobial peptide resistance module.
Nat Commun, 14, 2023
|
|
8G3A
 
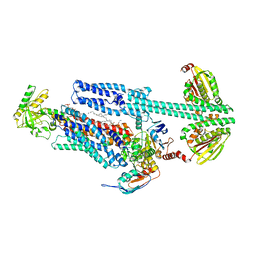 | | BceAB-S nucleotide free TM state 1 | | Descriptor: | Bacitracin export ATP-binding protein BceA, Bacitracin export permease protein BceB, OLEIC ACID, ... | | Authors: | George, N.L, Orlando, B.J. | | Deposit date: | 2023-02-07 | | Release date: | 2023-06-21 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Architecture of a complete Bce-type antimicrobial peptide resistance module.
Nat Commun, 14, 2023
|
|
8G4C
 
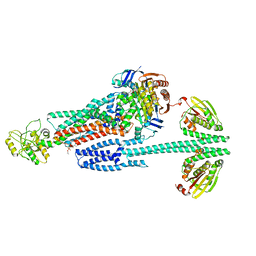 | | BceABS ATPgS high res TM | | Descriptor: | Bacitracin export ATP-binding protein BceA, Bacitracin export permease protein BceB, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | George, N.L, Orlando, B.J. | | Deposit date: | 2023-02-09 | | Release date: | 2023-06-21 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Architecture of a complete Bce-type antimicrobial peptide resistance module.
Nat Commun, 14, 2023
|
|
