2Y64
 
 | | Xylopentaose binding mutated (X-2 L110F) CBM4-2 Carbohydrate Binding Module from a Thermostable Rhodothermus marinus Xylanase | | Descriptor: | CALCIUM ION, XYLANASE, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose | | Authors: | von Schantz, L, Hakansson, M, Logan, D.T, Walse, B, Osterlin, J, Nordberg-Karlsson, E, Ohlin, M. | | Deposit date: | 2011-01-19 | | Release date: | 2012-03-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural basis for carbohydrate-binding specificity--a comparative assessment of two engineered carbohydrate-binding modules.
Glycobiology, 22, 2012
|
|
5DPN
 
 | | Engineered CBM X-2 L110F in complex with branched carbohydrate XXXG. | | Descriptor: | CALCIUM ION, Xylanase, alpha-D-xylopyranose-(1-6)-beta-D-glucopyranose-(1-4)-[alpha-D-xylopyranose-(1-6)]beta-D-glucopyranose-(1-4)-[alpha-D-xylopyranose-(1-6)]beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Ohlin, M. | | Deposit date: | 2015-09-13 | | Release date: | 2015-10-28 | | Last modified: | 2024-05-01 | | Method: | NEUTRON DIFFRACTION (1.6 Å), X-RAY DIFFRACTION | | Cite: | Neutron Crystallographic Studies Reveal Hydrogen Bond and Water-Mediated Interactions between a Carbohydrate-Binding Module and Its Bound Carbohydrate Ligand.
Biochemistry, 54, 2015
|
|
8XMZ
 
 | |
4MGQ
 
 | | PbXyn10C CBM APO | | Descriptor: | CALCIUM ION, Glycosyl hydrolase family 10 | | Authors: | Chekan, J.R, Nair, S.K. | | Deposit date: | 2013-08-28 | | Release date: | 2014-08-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Xylan utilization in human gut commensal bacteria is orchestrated by unique modular organization of polysaccharide-degrading enzymes.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3JXS
 
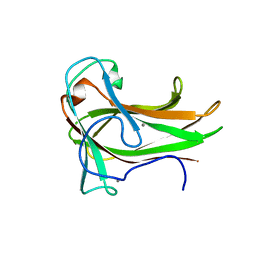 | | Crystal structure of XG34, an evolved xyloglucan binding CBM | | Descriptor: | ACETATE ION, CALCIUM ION, Xylanase | | Authors: | Divne, C, Tan, T.-C, Brumer, H, Gullfot, F. | | Deposit date: | 2009-09-21 | | Release date: | 2009-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The crystal structure of XG-34, an evolved xyloglucan-specific carbohydrate-binding module.
Proteins, 78, 2009
|
|
4BJ0
 
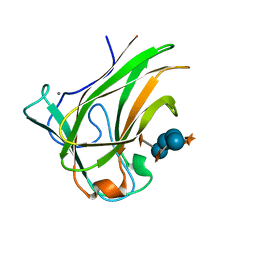 | | Xyloglucan binding module (CBM4-2 X2-L110F) in complex with branched xyloses | | Descriptor: | CALCIUM ION, XYLANASE, alpha-D-glucopyranose, ... | | Authors: | Schantz, L, Hakansson, M, Logan, D.T, Nordberg-Karlsson, E, Ohlin, M. | | Deposit date: | 2013-04-15 | | Release date: | 2014-04-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Carbohydrate Binding Module Recognition of Xyloglucan Defined by Polar Contacts with Branching Xyloses and Ch-Pi Interactions.
Proteins, 82, 2014
|
|
1DYO
 
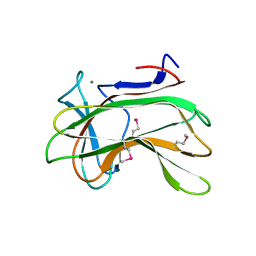 | | Xylan-Binding Domain from CBM 22, formally x6b domain | | Descriptor: | CALCIUM ION, ENDO-1,4-BETA-XYLANASE Y | | Authors: | Davies, G.J, Charnock, S.J, Gilbert, H.J, Fontes, C.M.G.A. | | Deposit date: | 2000-02-03 | | Release date: | 2000-07-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The X6 Thermostabilising Domains of Xylanases are Carbohydrate Binding Modules: Structure and Biochemistry of the Clostridium Thermocellum X6B Domain
Biochemistry, 39, 2000
|
|
1ULP
 
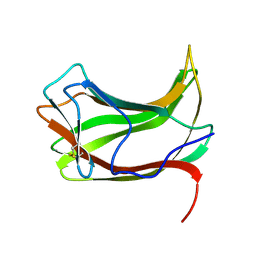 | |
1ULO
 
 | |
3OEA
 
 | | Crystal structure of the Q121E mutants of C.polysaccharolyticus CBM16-1 bound to cellopentaose | | Descriptor: | CALCIUM ION, S-layer associated multidomain endoglucanase, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Agarwal, V, Nair, S.K. | | Deposit date: | 2010-08-12 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Mutational insights into the roles of amino acid residues in ligand binding for two closely related family 16 carbohydrate binding modules.
J.Biol.Chem., 285, 2010
|
|
3OEB
 
 | |
3P6B
 
 | | The crystal structure of CelK CBM4 from Clostridium thermocellum | | Descriptor: | ACETATE ION, CALCIUM ION, Cellulose 1,4-beta-cellobiosidase, ... | | Authors: | Alahuhta, P.M, Luo, Y, Lunin, V.V. | | Deposit date: | 2010-10-11 | | Release date: | 2011-08-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of CBM4 from Clostridium thermocellum cellulase K.
Acta Crystallogr.,Sect.F, 67, 2011
|
|
1GUI
 
 | | CBM4 structure and function | | Descriptor: | CALCIUM ION, GLYCEROL, LAMINARINASE 16A, ... | | Authors: | Nurizzo, D, Notenboom, V, Davies, G.J. | | Deposit date: | 2002-01-27 | | Release date: | 2002-09-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Differential Oligosaccharide Recognition by Evolutionarily-Related Beta-1,4 and Beta-1,3 Glucan-Binding Modules
J.Mol.Biol., 319, 2002
|
|
1GU3
 
 | | CBM4 structure and function | | Descriptor: | ENDOGLUCANASE C, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Nurizzo, D, Notenboom, V, Davies, G.J. | | Deposit date: | 2002-01-22 | | Release date: | 2002-09-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Differential Oligosaccharide Recognition by Evolutionarily-Related Beta-1,4 and Beta-1,3 Glucan-Binding Modules
J.Mol.Biol., 319, 2002
|
|
1H6X
 
 | | The role of conserved amino acids in the cleft of the C-terminal family 22 carbohydrate binding module of Clostridium thermocellum Xyn10B in ligand binding | | Descriptor: | CALCIUM ION, ENDO-1,4-BETA-XYLANASE Y | | Authors: | Xie, H, Bolam, D.N, Charnock, S.J, Davies, G.J, Williamson, M.P, Simpson, P.J, Fontes, C.M.G.A, Ferreira, L.M.A, Gilbert, H.J. | | Deposit date: | 2001-06-29 | | Release date: | 2002-06-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Clostridium Thermocellum Xyn10B Carbohydrate-Binding Module 22-2: The Role of Conserved Amino Acids in Ligand Binding
Biochemistry, 40, 2001
|
|
1H6Y
 
 | | The role of conserved amino acids in the cleft of the C-terminal family 22 carbohydrate binding module of Clostridium thermocellum Xyn10B in ligand binding | | Descriptor: | CALCIUM ION, ENDO-1,4-BETA-XYLANASE Y | | Authors: | Xie, H, Bolam, D.N, Charnock, S.J, Davies, G.J, Williamson, M.P, Simpson, P.J, Fontes, C.M.G.A, Ferreira, L.M.A, Gilbert, H.J. | | Deposit date: | 2001-06-29 | | Release date: | 2002-06-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Clostridium Thermocellum Xyn10B Carbohydrate-Binding Module 22-2: The Role of Conserved Amino Acids in Ligand Binding
Biochemistry, 40, 2001
|
|
4XUR
 
 | |
4XUQ
 
 | |
4XUO
 
 | |
4XUT
 
 | | Structure of the CBM22-2 xylan-binding domain in complex with 1,3:1,4 Beta-glucotetraose B from Paenibacillus barcinonensis Xyn10C | | Descriptor: | CALCIUM ION, Endo-1,4-beta-xylanase C, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Sainz-Polo, M.A, Sanz-Aparicio, J. | | Deposit date: | 2015-01-26 | | Release date: | 2015-06-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Exploring Multimodularity in Plant Cell Wall Deconstruction: STRUCTURAL AND FUNCTIONAL ANALYSIS OF Xyn10C CONTAINING THE CBM22-1-CBM22-2 TANDEM.
J.Biol.Chem., 290, 2015
|
|
4XUP
 
 | |
4XUN
 
 | |
2Y6G
 
 | | Cellopentaose binding mutated (X-2 L110F) CBM4-2 Carbohydrate Binding Module from a Thermostable Rhodothermus marinus Xylanase | | Descriptor: | CALCIUM ION, XYLANASE, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | von Schantz, L, Hakansson, M, Logan, D.T, Walse, B, Osterlin, J, Nordberg-Karlsson, E, Ohlin, M. | | Deposit date: | 2011-01-21 | | Release date: | 2012-03-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural basis for carbohydrate-binding specificity--a comparative assessment of two engineered carbohydrate-binding modules.
Glycobiology, 22, 2012
|
|
2Y6L
 
 | | Xylopentaose binding X-2 engineered mutated CBM4-2 Carbohydrate Binding Module from a Thermostable Rhodothermus marinus Xylanase | | Descriptor: | CALCIUM ION, XYLANASE, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose | | Authors: | von Schantz, L, Hakansson, M, Logan, D.T, Walse, B, Osterlin, J, Nordberg-Karlsson, E, Ohlin, M. | | Deposit date: | 2011-01-24 | | Release date: | 2012-03-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Structural basis for carbohydrate-binding specificity--a comparative assessment of two engineered carbohydrate-binding modules.
Glycobiology, 22, 2012
|
|
2ZEX
 
 | | Family 16 carbohydrate binding module | | Descriptor: | CALCIUM ION, S-layer associated multidomain endoglucanase, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Bae, B, Nair, S.K. | | Deposit date: | 2007-12-18 | | Release date: | 2008-03-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Molecular Basis for the Selectivity and Specificity of Ligand Recognition by the Family 16 Carbohydrate-binding Modules from Thermoanaerobacterium polysaccharolyticum ManA
J.Biol.Chem., 283, 2008
|
|
