2OTN
 
 | | Crystal structure of the catalytically active form of diaminopimelate epimerase from Bacillus anthracis | | Descriptor: | Diaminopimelate epimerase | | Authors: | Matho, M.H, Fukuda, K, Santelli, E, Jaroszewski, L, Liddington, R.C, Roper, D. | | Deposit date: | 2007-02-08 | | Release date: | 2008-03-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure and inhibition of a catalytically active form of diaminopimelate epimerase (DapF)from Bacillus anthracis
To be Published
|
|
5M47
 
 | |
7VDY
 
 | | Crystal structure of O-ureidoserine racemase | | Descriptor: | O-ureido-serine racemase, SULFATE ION | | Authors: | Oda, K, Matoba, Y. | | Deposit date: | 2021-09-07 | | Release date: | 2021-12-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Crystal structure of O-ureidoserine racemase found in the d-cycloserine biosynthetic pathway.
Proteins, 90, 2022
|
|
1BWZ
 
 | |
3FVE
 
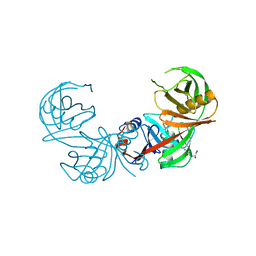 | | Crystal structure of diaminopimelate epimerase Mycobacterium tuberculosis DapF | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Diaminopimelate epimerase, GLYCEROL | | Authors: | Usha, V, Dover, L.G, Roper, D.I, Futterer, K, Besra, G.S. | | Deposit date: | 2009-01-15 | | Release date: | 2009-01-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the diaminopimelate epimerase DapF from Mycobacterium tuberculosis
Acta Crystallogr.,Sect.D, 65, 2009
|
|
1GQZ
 
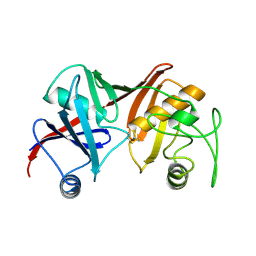 | |
8QZY
 
 | |
2Q9H
 
 | | Crystal structure of the C73S mutant of diaminopimelate epimerase | | Descriptor: | ACETIC ACID, Diaminopimelate epimerase, L(+)-TARTARIC ACID | | Authors: | Pillai, B, Cherney, M, Diaper, C.M, Sutherland, A, Blanchard, J.S, Vederas, J.C, James, M.N.G. | | Deposit date: | 2007-06-12 | | Release date: | 2007-10-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Dynamics of catalysis revealed from the crystal structures of mutants of diaminopimelate epimerase.
Biochem.Biophys.Res.Commun., 363, 2007
|
|
4IK0
 
 | | Crystal structure of diaminopimelate epimerase Y268A mutant from Escherichia coli | | Descriptor: | Diaminopimelate epimerase, IODIDE ION | | Authors: | Hor, L, Dobson, R.C.J, Hutton, C.A, Perugini, M.A. | | Deposit date: | 2012-12-24 | | Release date: | 2013-02-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Dimerization of bacterial diaminopimelate epimerase is essential for catalysis
J.Biol.Chem., 288, 2013
|
|
4IJZ
 
 | | Crystal structure of diaminopimelate epimerase from Escherichia coli | | Descriptor: | Diaminopimelate epimerase, NITRATE ION | | Authors: | Hor, L, Dobson, R.C.J, Hutton, C.A, Perugini, M.A. | | Deposit date: | 2012-12-24 | | Release date: | 2013-02-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Dimerization of bacterial diaminopimelate epimerase is essential for catalysis
J.Biol.Chem., 288, 2013
|
|
2GKE
 
 | | Crystal structure of diaminopimelate epimerase in complex with an irreversible inhibitor LL-AziDAP | | Descriptor: | (2S,6S)-2,6-DIAMINO-2-METHYLHEPTANEDIOIC ACID, ACETIC ACID, Diaminopimelate epimerase, ... | | Authors: | Pillai, B, Cherney, M.M, Diaper, C.M, Sutherland, A, Blanchard, J.S, Vederas, J.C, James, M.N. | | Deposit date: | 2006-04-01 | | Release date: | 2006-05-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural insights into stereochemical inversion by diaminopimelate epimerase: An antibacterial drug target.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
3EJX
 
 | | Crystal structure of diaminopimelate epimerase from Arabidopsis thaliana in complex with LL-AziDAP | | Descriptor: | (2S,6S)-2,6-DIAMINO-2-METHYLHEPTANEDIOIC ACID, Diaminopimelate epimerase, chloroplastic | | Authors: | Pillai, B, Moorthie, V.A, Cherney, M.M, Vederas, J.C, James, M.N.G. | | Deposit date: | 2008-09-18 | | Release date: | 2009-02-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of diaminopimelate epimerase from Arabidopsis thaliana, an amino acid racemase critical for L-lysine biosynthesis.
J.Mol.Biol., 385, 2009
|
|
3EKM
 
 | | Crystal structure of diaminopimelate epimerase form arabidopsis thaliana in complex with irreversible inhibitor DL-AziDAP | | Descriptor: | (2R,6S)-2,6-DIAMINO-2-METHYLHEPTANEDIOIC ACID, Diaminopimelate epimerase, chloroplastic | | Authors: | Pillai, B, Moorthie, V.A, Cherney, M.M, van Belkum, M.J, Vederas, J.C, James, M.N.G. | | Deposit date: | 2008-09-19 | | Release date: | 2009-02-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of diaminopimelate epimerase from Arabidopsis thaliana, an amino acid racemase critical for L-lysine biosynthesis.
J.Mol.Biol., 385, 2009
|
|
2Q9J
 
 | | Crystal structure of the C217S mutant of diaminopimelate epimerase | | Descriptor: | 1,2-ETHANEDIOL, Diaminopimelate epimerase, SULFATE ION | | Authors: | Pillai, B, Cherney, M, Diaper, C.M, Sutherland, A, Blanchard, J.S, Vederas, J.C. | | Deposit date: | 2007-06-12 | | Release date: | 2007-10-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Dynamics of catalysis revealed from the crystal structures of mutants of diaminopimelate epimerase.
Biochem.Biophys.Res.Commun., 363, 2007
|
|
2GKJ
 
 | | Crystal structure of diaminopimelate epimerase in complex with an irreversible inhibitor DL-AZIDAP | | Descriptor: | (2R,6S)-2,6-DIAMINO-2-METHYLHEPTANEDIOIC ACID, ACETIC ACID, Diaminopimelate epimerase | | Authors: | Pillai, B, Cherney, M.M, Diaper, C.M, Sutherland, A, Blanchard, J.S, Vederas, J.C, James, M.N. | | Deposit date: | 2006-04-02 | | Release date: | 2006-05-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insights into stereochemical inversion by diaminopimelate epimerase: An antibacterial drug target.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
5H2Y
 
 | |
5H2G
 
 | |
5HA4
 
 | |
5YGU
 
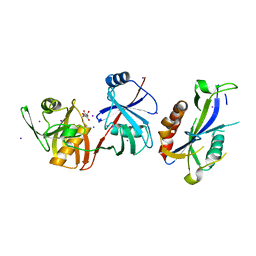 | | Crystal structure of Escherichia coli (strain K12) mRNA Decapping Complex RppH-DapF | | Descriptor: | Diaminopimelate epimerase, IODIDE ION, L(+)-TARTARIC ACID, ... | | Authors: | Wang, Q, Guan, Z.Y, Zhang, D.L, Zou, T.T, Yin, P. | | Deposit date: | 2017-09-27 | | Release date: | 2018-06-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.298 Å) | | Cite: | DapF stabilizes the substrate-favoring conformation of RppH to stimulate its RNA-pyrophosphohydrolase activity in Escherichia coli.
Nucleic Acids Res., 46, 2018
|
|
6D1V
 
 | |
6D13
 
 | | Crystal structure of E.coli RppH-DapF complex | | Descriptor: | CHLORIDE ION, Diaminopimelate epimerase, IODIDE ION, ... | | Authors: | Gao, A, Serganov, A. | | Deposit date: | 2018-04-11 | | Release date: | 2018-05-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | Structural and kinetic insights into stimulation of RppH-dependent RNA degradation by the metabolic enzyme DapF.
Nucleic Acids Res., 46, 2018
|
|
6D1Q
 
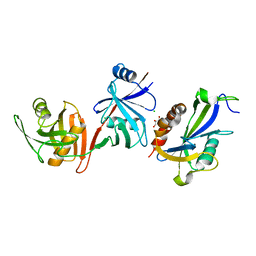 | | Crystal structure of E. coli RppH-DapF complex, monomer | | Descriptor: | CHLORIDE ION, Diaminopimelate epimerase, GLYCEROL, ... | | Authors: | Gao, A, Serganov, A. | | Deposit date: | 2018-04-12 | | Release date: | 2018-05-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural and kinetic insights into stimulation of RppH-dependent RNA degradation by the metabolic enzyme DapF.
Nucleic Acids Res., 46, 2018
|
|
6VCK
 
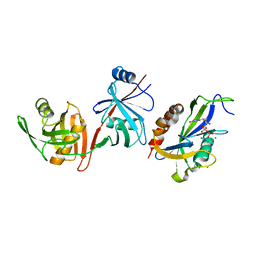 | | Crystal structure of E.coli RppH-DapF in complex with GDP, Mg2+ and F- | | Descriptor: | CHLORIDE ION, Diaminopimelate epimerase, FLUORIDE ION, ... | | Authors: | Gao, A, Vasilyev, N, Kaushik, A, Duan, W, Serganov, A. | | Deposit date: | 2019-12-21 | | Release date: | 2020-02-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Principles of RNA and nucleotide discrimination by the RNA processing enzyme RppH.
Nucleic Acids Res., 48, 2020
|
|
6VCM
 
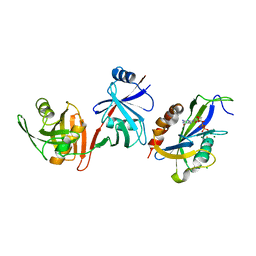 | | Crystal structure of E.coli RppH-DapF in complex with GTP, Mg2+ and F- | | Descriptor: | CHLORIDE ION, Diaminopimelate epimerase, FLUORIDE ION, ... | | Authors: | Gao, A, Vasilyev, N, Kaushik, A, Duan, W, Serganov, A. | | Deposit date: | 2019-12-21 | | Release date: | 2020-02-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Principles of RNA and nucleotide discrimination by the RNA processing enzyme RppH.
Nucleic Acids Res., 48, 2020
|
|
6VCL
 
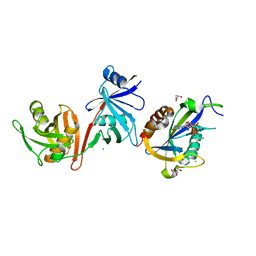 | | Crystal structure of E.coli RppH-DapF in complex with pppGpp, Mg2+ and F- | | Descriptor: | CHLORIDE ION, Diaminopimelate epimerase, FLUORIDE ION, ... | | Authors: | Gao, A, Vasilyev, N, Kaushik, A, Duan, W, Serganov, A. | | Deposit date: | 2019-12-21 | | Release date: | 2020-02-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Principles of RNA and nucleotide discrimination by the RNA processing enzyme RppH.
Nucleic Acids Res., 48, 2020
|
|
