3CQB
 
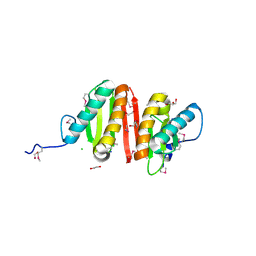 | | Crystal structure of heat shock protein HtpX domain from Vibrio parahaemolyticus RIMD 2210633 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Wu, R, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-04-02 | | Release date: | 2008-05-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | The crystal structure of heat shock protein HtpX domain from Vibrio parahaemolyticus RIMD 2210633.
To be Published
|
|
3C37
 
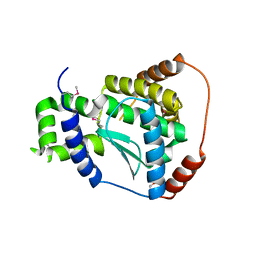 | | X-ray structure of the putative Zn-dependent peptidase Q74D82 at the resolution 1.7 A. Northeast Structural Genomics Consortium target GsR143A | | Descriptor: | DI(HYDROXYETHYL)ETHER, Peptidase, M48 family, ... | | Authors: | Kuzin, A.P, Chen, Y, Seetharaman, J, Vorobiev, S.M, Forouhar, F, Wang, D, Mao, L, Maglaqui, M, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-01-27 | | Release date: | 2008-02-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | X-ray structure of the putative Zn-dependent peptidase Q74D82 at the resolution 1.7 A.
To be Published
|
|
6SAR
 
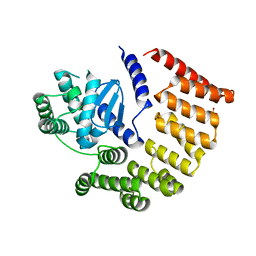 | | E coli BepA/YfgC | | Descriptor: | Beta-barrel assembly-enhancing protease, SULFATE ION, ZINC ION | | Authors: | Lovering, A.L, Cadby, I.T. | | Deposit date: | 2019-07-17 | | Release date: | 2020-08-26 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structure-Function Characterization of the Conserved Regulatory Mechanism of the Escherichia coli M48 Metalloprotease BepA.
J.Bacteriol., 203, 2020
|
|
4IL3
 
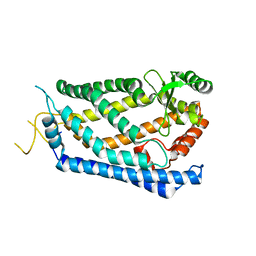 | | Crystal Structure of S. mikatae Ste24p | | Descriptor: | Ste24p, ZINC ION | | Authors: | Pryor Jr, E.E, Horanyi, P.S, Clark, K, Fedoriw, N, Connelly, S.M, Koszelak-Rosenblum, M, Zhu, G, Malkowski, M.G, Dumont, M.E, Wiener, M.C, Membrane Protein Structural Biology Consortium (MPSBC) | | Deposit date: | 2012-12-28 | | Release date: | 2013-02-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.102 Å) | | Cite: | Structure of the integral membrane protein CAAX protease Ste24p.
Science, 339, 2013
|
|
5SYT
 
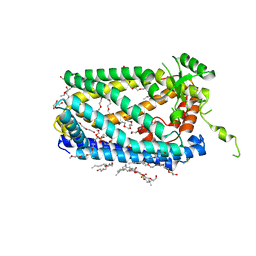 | | Crystal Structure of ZMPSTE24 | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CAAX prenyl protease 1 homolog, ... | | Authors: | Clark, K, Jenkins, J.L, Fedoriw, N, Dumont, M.E, Membrane Protein Structural Biology Consortium (MPSBC) | | Deposit date: | 2016-08-11 | | Release date: | 2016-11-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Human CaaX protease ZMPSTE24 expressed in yeast: Structure and inhibition by HIV protease inhibitors.
Protein Sci., 26, 2017
|
|
4AW6
 
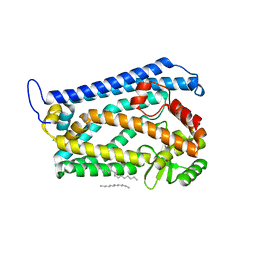 | | Crystal structure of the human nuclear membrane zinc metalloprotease ZMPSTE24 (FACE1) | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CAAX PRENYL PROTEASE 1 HOMOLOG, ZINC ION | | Authors: | Pike, A.C.W, Dong, Y.Y, Quigley, A, Dong, L, Cooper, C.D.O, Chaikuad, A, Goubin, S, Shrestha, L, Li, Q, Mukhopadhyay, S, Yang, J, Xia, X, Shintre, C.A, Barr, A.J, Berridge, G, Chalk, R, Bray, J.E, von Delft, F, Bullock, A, Bountra, C, Arrowsmith, C.H, Edwards, A, Burgess-Brown, N, Carpenter, E.P. | | Deposit date: | 2012-05-31 | | Release date: | 2012-07-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | The Structural Basis of Zmpste24-Dependent Laminopathies.
Science, 339, 2013
|
|
6AIT
 
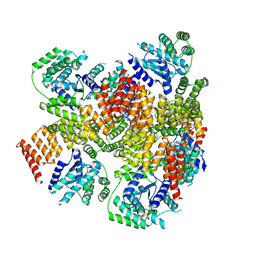 | | Crystal structure of E. coli BepA | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-barrel assembly-enhancing protease, ZINC ION | | Authors: | Umar, M.S.M, Tanaka, Y, Kamikubo, H, Tsukazaki, T. | | Deposit date: | 2018-08-24 | | Release date: | 2018-12-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.598 Å) | | Cite: | Structural Basis for the Function of the beta-Barrel Assembly-Enhancing Protease BepA.
J. Mol. Biol., 431, 2019
|
|
2YPT
 
 | | Crystal structure of the human nuclear membrane zinc metalloprotease ZMPSTE24 mutant (E336A) in complex with a synthetic CSIM tetrapeptide from the C-terminus of prelamin A | | Descriptor: | CAAX PRENYL PROTEASE 1 HOMOLOG, PRELAMIN-A/C, ZINC ION | | Authors: | Pike, A.C.W, Dong, Y.Y, Quigley, A, Dong, L, Savitsky, P, Cooper, C.D.O, Chaikuad, A, Goubin, S, Shrestha, L, Li, Q, Mukhopadhyay, S, Yang, J, Xia, X, Shintre, C.A, Barr, A.J, Berridge, G, Chalk, R, Bray, J.E, von Delft, F, Bullock, A, Bountra, C, Arrowsmith, C.H, Edwards, A, Burgess-Brown, N, Carpenter, E.P. | | Deposit date: | 2012-11-01 | | Release date: | 2012-12-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | The Structural Basis of Zmpste24-Dependent Laminopathies.
Science, 339, 2013
|
|
6BH8
 
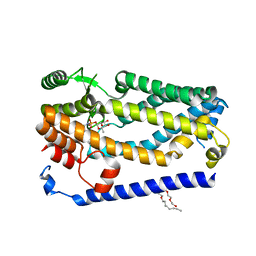 | | Crystal structure of ZMPSTE24 in complex with phosphoramidon | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, CAAX prenyl protease 1 homolog, N-ALPHA-L-RHAMNOPYRANOSYLOXY(HYDROXYPHOSPHINYL)-L-LEUCYL-L-TRYPTOPHAN, ... | | Authors: | Goblirsch, B.R, Arachea, B.T, Wiener, M.C. | | Deposit date: | 2017-10-30 | | Release date: | 2018-08-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.85 Å) | | Cite: | Phosphoramidon inhibits the integral membrane protein zinc metalloprotease ZMPSTE24.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
