3HSI
 
 | |
7V55
 
 | |
7V53
 
 | |
7SVP
 
 | |
6U8Z
 
 | |
6OHS
 
 | |
6OHM
 
 | |
7WDK
 
 | | The structure of PldA-PA3488 complex | | Descriptor: | Phospholipase D, Tli4_C domain-containing protein | | Authors: | Zhao, L, Yang, X.Y, Li, Z.Q. | | Deposit date: | 2021-12-21 | | Release date: | 2022-10-26 | | Last modified: | 2022-11-09 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Structural insights into PA3488-mediated inactivation of Pseudomonas aeruginosa PldA
Nat Commun, 13, 2022
|
|
6OHO
 
 | | Structure of human Phospholipase D2 catalytic domain | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, GLYCEROL, Phospholipase D2, ... | | Authors: | Metrick, C.M, Chodaparambil, J.V. | | Deposit date: | 2019-04-06 | | Release date: | 2020-02-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Human PLD structures enable drug design and characterization of isoenzyme selectivity.
Nat.Chem.Biol., 16, 2020
|
|
6OHP
 
 | |
6OHQ
 
 | |
6OHR
 
 | |
6KZ8
 
 | | Crystal structure of plant Phospholipase D alpha complex with phosphatidic acid | | Descriptor: | 1,2-DIOCTANOYL-SN-GLYCERO-3-PHOSPHATE, CALCIUM ION, Phospholipase D alpha 1 | | Authors: | Li, J.X, Yu, F, Zhang, P. | | Deposit date: | 2019-09-23 | | Release date: | 2019-11-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.291 Å) | | Cite: | Crystal structure of plant PLD alpha 1 reveals catalytic and regulatory mechanisms of eukaryotic phospholipase D.
Cell Res., 30, 2020
|
|
6KZ9
 
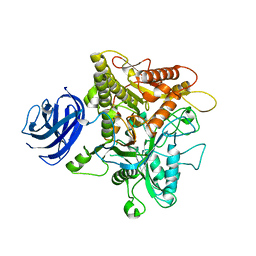 | | Crystal structure of plant Phospholipase D alpha | | Descriptor: | CALCIUM ION, Phospholipase D alpha 1 | | Authors: | Li, J.X, Yu, F, Zhang, P. | | Deposit date: | 2019-09-23 | | Release date: | 2019-10-30 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Crystal structure of plant PLD alpha 1 reveals catalytic and regulatory mechanisms of eukaryotic phospholipase D.
Cell Res., 30, 2020
|
|
