6SWY
 
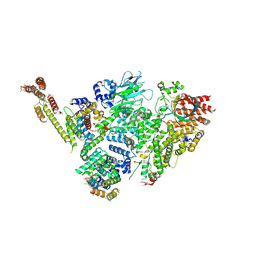 | | Structure of active GID E3 ubiquitin ligase complex minus Gid2 and delta Gid9 RING domain | | Descriptor: | Glucose-induced degradation protein 8, Protein FYV10,Protein FYV10,Protein FYV10,Protein FYV10,Protein FYV10,Protein FYV10,Protein FYV10, Vacuolar import and degradation protein 24, ... | | Authors: | Qiao, S, Prabu, J.R, Schulman, B.A. | | Deposit date: | 2019-09-24 | | Release date: | 2019-11-20 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Interconversion between Anticipatory and Active GID E3 Ubiquitin Ligase Conformations via Metabolically Driven Substrate Receptor Assembly
Mol.Cell, 77, 2020
|
|
7WUG
 
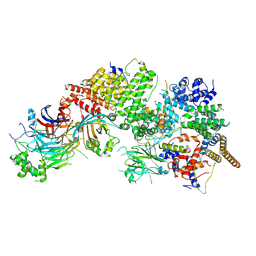 | | GID subcomplex: Gid12 bound Substrate Receptor Scaffolding module | | Descriptor: | Glucose-induced degradation protein 8, HLJ1_G0042170.mRNA.1.CDS.1, Vacuolar import and degradation protein 24, ... | | Authors: | Qiao, S, Cheng, J.D, Schulman, B.A. | | Deposit date: | 2022-02-08 | | Release date: | 2022-06-08 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures of Gid12-bound GID E3 reveal steric blockade as a mechanism inhibiting substrate ubiquitylation.
Nat Commun, 13, 2022
|
|
7NSB
 
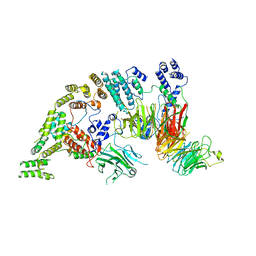 | | Supramolecular assembly module of yeast Chelator-GID SR4 E3 ubiquitin ligase | | Descriptor: | Glucose-induced degradation protein 7, Glucose-induced degradation protein 8, Vacuolar import and degradation protein 30 | | Authors: | Chrustowicz, J, Sherpa, D, Prabu, J.R, Schulman, B.A. | | Deposit date: | 2021-03-05 | | Release date: | 2021-05-05 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | GID E3 ligase supramolecular chelate assembly configures multipronged ubiquitin targeting of an oligomeric metabolic enzyme.
Mol.Cell, 81, 2021
|
|
