2BAY
 
 | | Crystal structure of the Prp19 U-box dimer | | Descriptor: | Pre-mRNA splicing factor PRP19 | | Authors: | Vander Kooi, C.W, Ohi, M.D, Rosenberg, J.A, Oldham, M.L, Newcomer, M.E, Gould, K.L, Chazin, W.J. | | Deposit date: | 2005-10-15 | | Release date: | 2006-01-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Prp19 U-box Crystal Structure Suggests a Common Dimeric Architecture for a Class of Oligomeric E3 Ubiquitin Ligases.
Biochemistry, 45, 2006
|
|
5M8C
 
 | | Spliceosome component | | Descriptor: | Pre-mRNA-processing factor 19 | | Authors: | Moura, T.R, Pena, V. | | Deposit date: | 2016-10-28 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Prp19/Pso4 Is an Autoinhibited Ubiquitin Ligase Activated by Stepwise Assembly of Three Splicing Factors.
Mol. Cell, 69, 2018
|
|
4ZB4
 
 | | Spliceosome component | | Descriptor: | Pre-mRNA-processing factor 19 | | Authors: | Rocha de Moura, T, Pena, P. | | Deposit date: | 2015-04-14 | | Release date: | 2016-09-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Spliceosome component
To Be Published
|
|
3LRV
 
 | |
6J6G
 
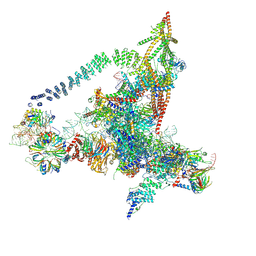 | | Cryo-EM structure of the yeast B*-a2 complex at an average resolution of 3.2 angstrom | | Descriptor: | ACT1 pre-mRNA, GUANOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Wan, R, Bai, R, Yan, C, Lei, J, Shi, Y. | | Deposit date: | 2019-01-15 | | Release date: | 2019-04-24 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structures of the Catalytically Activated Yeast Spliceosome Reveal the Mechanism of Branching.
Cell, 177, 2019
|
|
6BK8
 
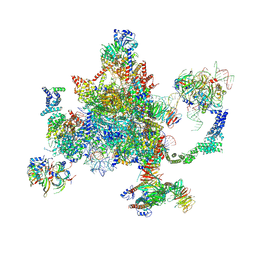 | | S. cerevisiae spliceosomal post-catalytic P complex | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, Lea1, ... | | Authors: | Liu, S, Li, X, Zhou, Z.H, Zhao, R. | | Deposit date: | 2017-11-07 | | Release date: | 2018-02-21 | | Last modified: | 2020-10-14 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of the yeast spliceosomal postcatalytic P complex.
Science, 358, 2017
|
|
5GMK
 
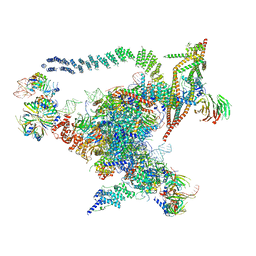 | | Cryo-EM structure of the Catalytic Step I spliceosome (C complex) at 3.4 angstrom resolution | | Descriptor: | 5'-Exon, 5'-Splicing Site, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Wan, R, Yan, C, Bai, R, Huang, G, Shi, Y. | | Deposit date: | 2016-07-14 | | Release date: | 2016-08-17 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of a yeast catalytic step I spliceosome at 3.4 angstrom resolution
Science, 353, 2016
|
|
5Y88
 
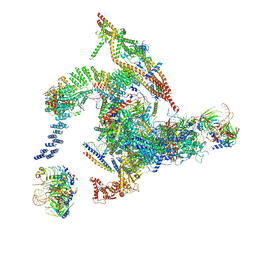 | | Cryo-EM structure of the intron-lariat spliceosome ready for disassembly from S.cerevisiae at 3.5 angstrom | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, Intron lariat, ... | | Authors: | Wan, R, Yan, C, Bai, R, Lei, J, Shi, Y. | | Deposit date: | 2017-08-20 | | Release date: | 2018-08-01 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Structure of an Intron Lariat Spliceosome from Saccharomyces cerevisiae
Cell(Cambridge,Mass.), 171, 2017
|
|
5GM6
 
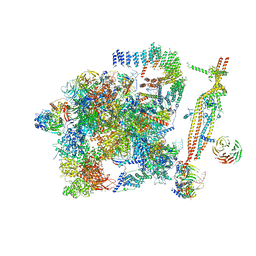 | | Cryo-EM structure of the activated spliceosome (Bact complex) at 3.5 angstrom resolution | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cold sensitive U2 snRNA suppressor 1, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Yan, C, Wan, R, Bai, R, Huang, G, Shi, Y. | | Deposit date: | 2016-07-12 | | Release date: | 2016-09-21 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of a yeast activated spliceosome at 3.5 angstrom resolution
Science, 353, 2016
|
|
5YLZ
 
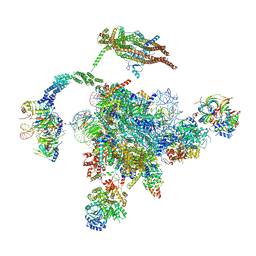 | | Cryo-EM Structure of the Post-catalytic Spliceosome from Saccharomyces cerevisiae at 3.6 angstrom | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Wan, R, Yan, C, Bai, R, Lei, J, Shi, Y. | | Deposit date: | 2017-10-20 | | Release date: | 2018-07-18 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure of the Post-catalytic Spliceosome from Saccharomyces cerevisiae
Cell, 171, 2017
|
|
6J6H
 
 | | Cryo-EM structure of the yeast B*-a1 complex at an average resolution of 3.6 angstrom | | Descriptor: | ACT1 pre-mRNA, GUANOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Wan, R, Bai, R, Yan, C, Lei, J, Shi, Y. | | Deposit date: | 2019-01-15 | | Release date: | 2019-04-24 | | Last modified: | 2020-10-14 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structures of the Catalytically Activated Yeast Spliceosome Reveal the Mechanism of Branching.
Cell, 177, 2019
|
|
6EXN
 
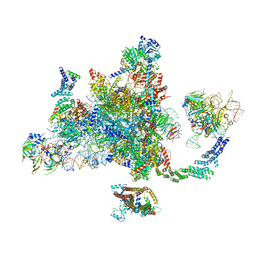 | | Post-catalytic P complex spliceosome with 3' splice site docked | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, Intron lariat: UBC4 RNA, ... | | Authors: | Wilkinson, M.E, Fica, S.M, Galej, W.P, Norman, C.M, Newman, A.J, Nagai, K. | | Deposit date: | 2017-11-08 | | Release date: | 2018-01-17 | | Last modified: | 2020-10-07 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Postcatalytic spliceosome structure reveals mechanism of 3'-splice site selection.
Science, 358, 2017
|
|
6J6Q
 
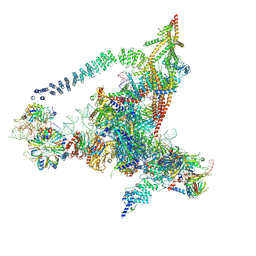 | | Cryo-EM structure of the yeast B*-b2 complex at an average resolution of 3.7 angstrom | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Wan, R, Bai, R, Yan, C, Lei, J, Shi, Y. | | Deposit date: | 2019-01-15 | | Release date: | 2019-04-24 | | Last modified: | 2020-10-14 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structures of the Catalytically Activated Yeast Spliceosome Reveal the Mechanism of Branching.
Cell, 177, 2019
|
|
5LJ5
 
 | | Overall structure of the yeast spliceosome immediately after branching. | | Descriptor: | CWC15, CWC22, Exon 1 (5' exon) of UBC4 pre-mRNA, ... | | Authors: | Galej, W.P, Wilkinson, M.F, Fica, S.M, Oubridge, C, Newman, A.J, Nagai, K. | | Deposit date: | 2016-07-17 | | Release date: | 2016-08-31 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structure of the spliceosome immediately after branching.
Nature, 537, 2016
|
|
6J6N
 
 | | Cryo-EM structure of the yeast B*-b1 complex at an average resolution of 3.86 angstrom | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Wan, R, Bai, R, Yan, C, Lei, J, Shi, Y. | | Deposit date: | 2019-01-15 | | Release date: | 2019-04-24 | | Last modified: | 2020-10-14 | | Method: | ELECTRON MICROSCOPY (3.86 Å) | | Cite: | Structures of the Catalytically Activated Yeast Spliceosome Reveal the Mechanism of Branching.
Cell, 177, 2019
|
|
5WSG
 
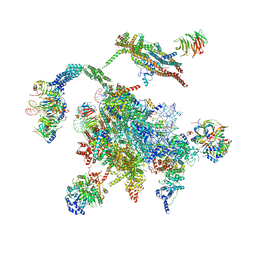 | | Cryo-EM structure of the Catalytic Step II spliceosome (C* complex) at 4.0 angstrom resolution | | Descriptor: | 3'-exon-intron, 3'-intron-lariat, 5'-exon, ... | | Authors: | Yan, C, Wan, R, Bai, R, Huang, G, Shi, Y. | | Deposit date: | 2016-12-07 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure of a yeast step II catalytically activated spliceosome
Science, 355, 2017
|
|
5MQ0
 
 | | Structure of a spliceosome remodeled for exon ligation | | Descriptor: | 3'-EXON OF UBC4 PRE-MRNA, BOUND BY PRP22 HELICASE, 5'-EXON OF UBC4 PRE-MRNA, ... | | Authors: | Fica, S.M, Oubridge, C, Galej, W.P, Wilkinson, M.E, Newman, A.J, Bai, X.-C, Nagai, K. | | Deposit date: | 2016-12-19 | | Release date: | 2017-01-18 | | Last modified: | 2020-10-07 | | Method: | ELECTRON MICROSCOPY (4.17 Å) | | Cite: | Structure of a spliceosome remodelled for exon ligation.
Nature, 542, 2017
|
|
1N87
 
 | |
