3VZ6
 
 | |
1KID
 
 | |
3VZ7
 
 | |
3VZ8
 
 | |
1SX3
 
 | | GroEL14-(ATPgammaS)14 | | Descriptor: | MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, POTASSIUM ION, ... | | Authors: | Chaudhry, C, Horwich, A.L, Brunger, A.T, Adams, P.D. | | Deposit date: | 2004-03-30 | | Release date: | 2005-03-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Exploring the structural dynamics of the E.coli chaperonin GroEL using translation-libration-screw crystallographic refinement of intermediate states.
J.Mol.Biol., 342, 2004
|
|
1KP8
 
 | |
1DK7
 
 | |
1LA1
 
 | | Gro-EL Fragment (Apical Domain) Comprising Residues 188-379 | | Descriptor: | GroEL | | Authors: | Ashcroft, A.E, Brinker, A, Coyle, J.E, Weber, F, Kaiser, M, Moroder, L, Parsons, M.R, Jager, J, Hartl, U.F, Hayer-Hartl, M, Radford, S.E. | | Deposit date: | 2002-03-27 | | Release date: | 2002-04-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structural plasticity and noncovalent substrate binding in the GroEL apical domain. A study using electrospay ionization mass spectrometry and fluorescence binding studies.
J.Biol.Chem., 277, 2002
|
|
1DKD
 
 | |
1FYA
 
 | |
1FY9
 
 | |
8BL2
 
 | | Structure of GroEL-ATP complex plunge frozen 200 ms after reaction initiation | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Chaperonin GroEL, MAGNESIUM ION, ... | | Authors: | Dhurandhar, M, Torino, S, Efremov, R. | | Deposit date: | 2022-11-09 | | Release date: | 2023-08-09 | | Last modified: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Time-resolved cryo-EM using a combination of droplet microfluidics with on-demand jetting.
Nat.Methods, 20, 2023
|
|
8BKZ
 
 | | GroEL:GroES-ATP complex under continuous turnover conditions | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Chaperonin GroEL, Co-chaperonin GroES, ... | | Authors: | Dhurandhar, M, Torino, S, Efremov, R. | | Deposit date: | 2022-11-09 | | Release date: | 2023-08-09 | | Last modified: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Time-resolved cryo-EM using a combination of droplet microfluidics with on-demand jetting.
Nat.Methods, 20, 2023
|
|
8S32
 
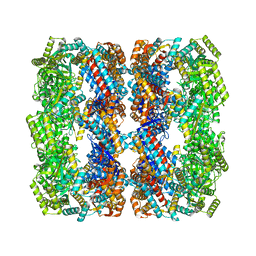 | | GroEL with bound GroTAC peptide | | Descriptor: | Chaperonin GroEL, GroTAC | | Authors: | Wroblewski, K, Izert-Nowakowska, M.A, Goral, T.K, Klimecka, M.M, Kmiecik, S, Gorna, M.W. | | Deposit date: | 2024-02-19 | | Release date: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (2.45 Å) | | Cite: | Depletion of essential GroEL protein in Escherichia coli using Clp-Interacting Peptidic Protein Erasers (CLIPPERs)
To Be Published
|
|
1JON
 
 | |
8P4M
 
 | | CryoEM structure of a C7-symmetrical GroEL7-GroES7 cage in presence of ADP-BeFx | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Chaperonin GroEL, ... | | Authors: | Wagner, J, Beck, F, Bracher, A, Caravajal, A.I, Wan, W, Bohn, S, Koerner, R, Baumeister, W, Fernandez-Busnadiego, R, Hartl, F.U. | | Deposit date: | 2023-05-23 | | Release date: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Visualizing chaperonin function in situ by cryo-electron tomography
Nature, 2024
|
|
8BMT
 
 | | Structure of GroEL:GroES-ATP complex plunge frozen 200 ms after reaction initiation | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Chaperonin GroEL, Co-chaperonin GroES, ... | | Authors: | Dhurandhar, M, Efremov, R, Torino, S. | | Deposit date: | 2022-11-10 | | Release date: | 2023-08-09 | | Last modified: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Time-resolved cryo-EM using a combination of droplet microfluidics with on-demand jetting.
Nat.Methods, 20, 2023
|
|
7XOK
 
 | |
8BM1
 
 | | Structure of GroEL:GroES-ATP complex under continuous turnover conditions | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Chaperonin GroEL, Co-chaperonin GroES, ... | | Authors: | Dhurandhar, M, Torino, S, Efremov, R. | | Deposit date: | 2022-11-10 | | Release date: | 2023-08-09 | | Last modified: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Time-resolved cryo-EM using a combination of droplet microfluidics with on-demand jetting.
Nat.Methods, 20, 2023
|
|
8BLC
 
 | | Structure of the GroEL-ATP complex plunge-frozen 50 ms after mixing with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Chaperonin GroEL, MAGNESIUM ION, ... | | Authors: | Dhurandhar, M, Torino, S, Efremov, R. | | Deposit date: | 2022-11-09 | | Release date: | 2023-08-09 | | Last modified: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Time-resolved cryo-EM using a combination of droplet microfluidics with on-demand jetting.
Nat.Methods, 20, 2023
|
|
1SS8
 
 | | GroEL | | Descriptor: | groEL protein | | Authors: | Chaudhry, C, Horwich, A.L, Brunger, A.T, Adams, P.D. | | Deposit date: | 2004-03-23 | | Release date: | 2005-03-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Exploring the structural dynamics of the E.coli chaperonin GroEL using translation-libration-screw crystallographic refinement of intermediate states.
J.Mol.Biol., 342, 2004
|
|
7XOJ
 
 | |
1OEL
 
 | |
8BMD
 
 | | Structure of GroEL-ATP complex under continuous turnover conditions | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Chaperonin GroEL, MAGNESIUM ION, ... | | Authors: | Dhurandhar, M, Torino, S, Efremov, R. | | Deposit date: | 2022-11-10 | | Release date: | 2023-08-09 | | Last modified: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Time-resolved cryo-EM using a combination of droplet microfluidics with on-demand jetting.
Nat.Methods, 20, 2023
|
|
1GRL
 
 | | THE CRYSTAL STRUCTURE OF THE BACTERIAL CHAPERONIN GROEL AT 2.8 ANGSTROMS | | Descriptor: | GROEL (HSP60 CLASS) | | Authors: | Braig, K, Otwinowski, Z, Hegde, R, Boisvert, D.C, Joachimiak, A, Horwich, A.L, Sigler, P.B. | | Deposit date: | 1995-03-07 | | Release date: | 1995-10-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The crystal structure of the bacterial chaperonin GroEL at 2.8 A.
Nature, 371, 1994
|
|
