2NLR
 
 | |
5GM5
 
 | | Crystal structure of FI-CMCase from Aspergillus aculeatus F-50 in complex with cellobiose | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Endoglucanase-1, SULFATE ION, ... | | Authors: | Huang, J.W, Liu, W.D, Zheng, Y.Y, Chen, C.C, Guo, R.T. | | Deposit date: | 2016-07-12 | | Release date: | 2017-05-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Crystal structure and genetic modifications of FI-CMCase from Aspergillus aculeatus F-50
Biochem. Biophys. Res. Commun., 478, 2016
|
|
5GM4
 
 | | Crystal structure of FI-CMCase from Aspergillus aculeatus F-50 in complex with cellotetrose | | Descriptor: | Endoglucanase-1, SULFATE ION, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Huang, J.W, Liu, W.D, Zheng, Y.Y, Chen, C.C, Guo, R.T. | | Deposit date: | 2016-07-12 | | Release date: | 2017-05-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal structure and genetic modifications of FI-CMCase from Aspergillus aculeatus F-50
Biochem. Biophys. Res. Commun., 478, 2016
|
|
5GM3
 
 | | Crystal structure of FI-CMCase from Aspergillus aculeatus F-50 | | Descriptor: | CACODYLATE ION, Endoglucanase-1, ZINC ION | | Authors: | Huang, J.W, Liu, W.D, Zheng, Y.Y, Chen, C.C, Guo, R.T. | | Deposit date: | 2016-07-12 | | Release date: | 2017-05-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Crystal structure and genetic modifications of FI-CMCase from Aspergillus aculeatus F-50
Biochem. Biophys. Res. Commun., 478, 2016
|
|
1CLC
 
 | |
7VT7
 
 | | Crystal structure of CBM deleted MtGlu5 in complex with CBI | | Descriptor: | Endoglucanase H, GLYCEROL, TRYPTOPHAN, ... | | Authors: | Ye, T.J, Ko, P.T, Huang, K.F, Wu, S.H. | | Deposit date: | 2021-10-28 | | Release date: | 2022-09-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Synergic action of an inserted carbohydrate-binding module in a glycoside hydrolase family 5 endoglucanase.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7VT8
 
 | | Crystal structure of MtGlu5 from Meiothermus taiwanensis WR-220 | | Descriptor: | Endoglucanase H, SULFATE ION, beta-D-glucopyranose | | Authors: | Ye, T.J, Ko, P.T, Huang, K.F, Wu, S.H. | | Deposit date: | 2021-10-28 | | Release date: | 2022-09-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Synergic action of an inserted carbohydrate-binding module in a glycoside hydrolase family 5 endoglucanase.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7VT4
 
 | | Crystal structure of mutant E393Q of MtGlu5 | | Descriptor: | Endoglucanase H, GLYCEROL, SULFATE ION | | Authors: | Ye, T.J, Ko, P.T, Huang, K.F, Wu, S.H. | | Deposit date: | 2021-10-28 | | Release date: | 2022-09-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Synergic action of an inserted carbohydrate-binding module in a glycoside hydrolase family 5 endoglucanase.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7VT6
 
 | | Crystal structure of CBM deleted MtGlu5 in complex with BGC. | | Descriptor: | Endoglucanase H, GLYCEROL, TRYPTOPHAN, ... | | Authors: | Ye, T.J, Ko, P.T, Huang, K.F, Wu, S.H. | | Deposit date: | 2021-10-28 | | Release date: | 2022-09-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Synergic action of an inserted carbohydrate-binding module in a glycoside hydrolase family 5 endoglucanase.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7VT5
 
 | | Crystal structure of CBM deleted MtGlu5 from Meiothermus taiwanensis WR-220 | | Descriptor: | Endoglucanase H, METHIONINE, TRYPTOPHAN | | Authors: | Ye, T.J, Ko, P.T, Huang, K.F, Wu, S.H. | | Deposit date: | 2021-10-28 | | Release date: | 2022-09-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Synergic action of an inserted carbohydrate-binding module in a glycoside hydrolase family 5 endoglucanase.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
1WZZ
 
 | | Structure of endo-beta-1,4-glucanase CMCax from Acetobacter xylinum | | Descriptor: | Probable endoglucanase, SULFATE ION | | Authors: | Yasutake, Y, Kawano, S, Tajima, K, Yao, M, Satoh, Y, Munekata, M, Tanaka, I, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-03-10 | | Release date: | 2006-03-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural characterization of the Acetobacter xylinum endo-beta-1,4-glucanase CMCax required for cellulose biosynthesis.
Proteins, 64, 2006
|
|
4L48
 
 | |
1EGZ
 
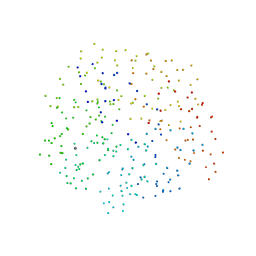 | | CELLULASE CEL5 FROM ERWINIA CHRYSANTHEMI, A FAMILY GH 5-2 ENZYME | | Descriptor: | CALCIUM ION, ENDOGLUCANASE Z | | Authors: | Czjzek, M, El Hassouni, M, Py, B, Barras, F. | | Deposit date: | 1999-03-18 | | Release date: | 1999-03-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Type II protein secretion in gram-negative pathogenic bacteria: the study of the structure/secretion relationships of the cellulase Cel5 (formerly EGZ) from Erwinia chrysanthemi
J.Mol.Biol., 310, 2001
|
|
4W65
 
 | |
7S8K
 
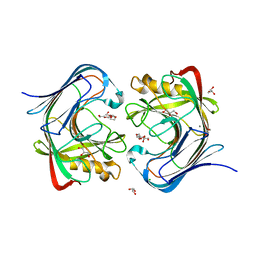 | | Crystal structure of a GH12-2 family cellulase from Thermococcus sp. 2319x1 | | Descriptor: | 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Stogios, P.J, Skarina, T, Khusnutdinova, A, Yakunin, A.F, Savchenko, A. | | Deposit date: | 2021-09-18 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | GH12-2 family cellulase
To Be Published
|
|
3WQ7
 
 | |
6A3H
 
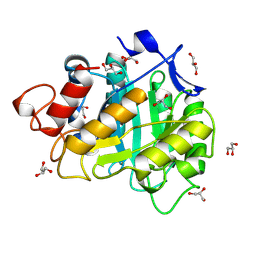 | | 2-DEOXY-2-FLURO-B-D-CELLOTRIOSYL/ENZYME INTERMEDIATE COMPLEX OF THE ENDOGLUCANASE CEL5A FROM BACILLUS AGARADHEARANS AT 1.6 ANGSTROM RESOLUTION | | Descriptor: | ENDOGLUCANASE, GLYCEROL, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-2-deoxy-2-fluoro-alpha-D-glucopyranose | | Authors: | Davies, G.J, Varrot, A, Dauter, M, Brzozowski, A.M, Schulein, M, Mackenzie, L, Withers, S.G. | | Deposit date: | 1998-07-22 | | Release date: | 1999-07-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Snapshots along an enzymatic reaction coordinate: analysis of a retaining beta-glycoside hydrolase.
Biochemistry, 37, 1998
|
|
6WQY
 
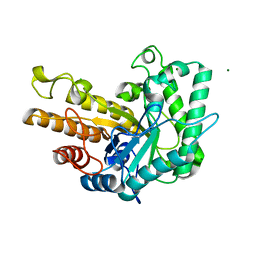 | | GH5-4 broad specificity endoglucanase from Bacteroides salanitronis | | Descriptor: | CHLORIDE ION, Cellulase, MAGNESIUM ION | | Authors: | Bingman, C.A, Smith, R.W, Glasgow, E.M, Fox, B.G. | | Deposit date: | 2020-04-29 | | Release date: | 2020-11-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | A structural and kinetic survey of GH5_4 endoglucanases reveals determinants of broad substrate specificity and opportunities for biomass hydrolysis.
J.Biol.Chem., 295, 2020
|
|
6FAO
 
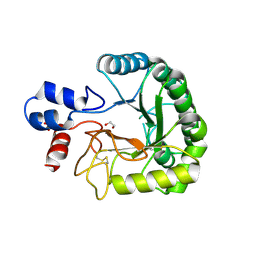 | | Discovery and characterization of a thermostable GH6 endoglucanase from a compost metagenome | | Descriptor: | 1,2-ETHANEDIOL, Glycoside hydrolase family 6, SULFATE ION | | Authors: | Jensen, M.S, Fredriksen, L, MacKenzie, A.K, Pope, P.B, Chylenski, P, Leiros, I, Williamson, A.K, Christopeit, T, Ostby, H, Vaaje-Kolstad, G, Eijsink, V.G.H. | | Deposit date: | 2017-12-15 | | Release date: | 2018-06-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Discovery and characterization of a thermostable two-domain GH6 endoglucanase from a compost metagenome.
PLoS ONE, 13, 2018
|
|
5IHS
 
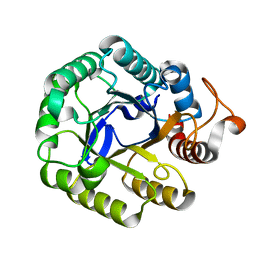 | | Structure of CHU_2103 from Cytophaga hutchinsonii | | Descriptor: | Endoglucanase, glycoside hydrolase family 5 protein | | Authors: | Silvaggi, N.R, Han, L. | | Deposit date: | 2016-02-29 | | Release date: | 2016-06-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Periplasmic Cytophaga hutchinsonii Endoglucanases Are Required for Use of Crystalline Cellulose as the Sole Source of Carbon and Energy.
Appl.Environ.Microbiol., 82, 2016
|
|
8OZ1
 
 | | CjCel5D endo-xyloglucanase bounc to CB665 covalent inhibitor | | Descriptor: | (1R,2S,3S,4S,5R,6R)-6-(HYDROXYMETHYL)CYCLOHEXANE-1,2,3,4,5-PENTOL, BORIC ACID, Cellulase, ... | | Authors: | McGregor, N.G.S, de Boer, C, Overkleeft, H.S, Davies, G.J. | | Deposit date: | 2023-05-06 | | Release date: | 2024-02-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | A Multiplexing Activity-Based Protein-Profiling Platform for Dissection of a Native Bacterial Xyloglucan-Degrading System.
Acs Cent.Sci., 9, 2023
|
|
8S3L
 
 | | X-ray crystal structure of LsAA9A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, COPPER (II) ION, ... | | Authors: | Frandsen, K.E.H, Di Domenico, V, Lo Leggio, L. | | Deposit date: | 2024-02-20 | | Release date: | 2025-03-05 | | Last modified: | 2025-09-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Anions and citrate inhibit LsAA9A, a lytic polysaccharide monooxygenase (LPMO).
Febs J., 292, 2025
|
|
8S3F
 
 | | X-ray crystal structure of LsAA9A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CITRIC ACID, COPPER (II) ION, ... | | Authors: | Frandsen, K.E.H, Di Domenico, V, Lo Leggio, L. | | Deposit date: | 2024-02-20 | | Release date: | 2025-03-05 | | Last modified: | 2025-09-24 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Anions and citrate inhibit LsAA9A, a lytic polysaccharide monooxygenase (LPMO).
Febs J., 292, 2025
|
|
7M1R
 
 | |
9B87
 
 | | Tetrameric cryo-EM structure of E. coli BcsZ | | Descriptor: | Endoglucanase | | Authors: | Verma, P, Zimmer, J. | | Deposit date: | 2024-03-29 | | Release date: | 2025-02-19 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (2.65 Å) | | Cite: | Insights into phosphoethanolamine cellulose synthesis and secretion across the Gram-negative cell envelope.
Nat Commun, 15, 2024
|
|
