1PUP
 
 | |
1PUQ
 
 | | Solution Structure of the MutT Pyrophosphohydrolase Complexed with Mg(2+) and 8-oxo-dGMP, a Tightly-bound Product | | Descriptor: | 8-OXO-2'-DEOXY-GUANOSINE-5'-MONOPHOSPHATE, MAGNESIUM ION, Mutator mutT protein | | Authors: | Massiah, M.A, Saraswat, V, Azurmendi, H.F, Mildvan, A.S. | | Deposit date: | 2003-06-25 | | Release date: | 2003-08-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure and NH exchange studies of the MutT pyrophosphohydrolase complexed with Mg(2+) and 8-oxo-dGMP, a tightly bound product.
Biochemistry, 42, 2003
|
|
1PUS
 
 | | Solution Structure of the MutT Pyrophosphohydrolase Complexed with Mg(2+) and 8-oxo-dGMP, a Tightly-bound Product | | Descriptor: | 8-OXO-2'-DEOXY-GUANOSINE-5'-MONOPHOSPHATE, MAGNESIUM ION, Mutator mutT protein | | Authors: | Massiah, M.A, Saraswat, V, Azurmendi, H.F, Mildvan, A.S. | | Deposit date: | 2003-06-25 | | Release date: | 2003-08-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure and NH exchange studies of the MutT pyrophosphohydrolase complexed with Mg(2+) and 8-oxo-dGMP, a tightly bound product.
Biochemistry, 42, 2003
|
|
1PUT
 
 | | AN NMR-DERIVED MODEL FOR THE SOLUTION STRUCTURE OF OXIDIZED PUTIDAREDOXIN, A 2FE, 2-S FERREDOXIN FROM PSEUDOMONAS | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, PUTIDAREDOXIN | | Authors: | Pochapsky, T.C, Ye, X.M, Ratnaswamy, G, Lyons, T.A. | | Deposit date: | 1994-07-09 | | Release date: | 1994-09-30 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | An NMR-derived model for the solution structure of oxidized putidaredoxin, a 2-Fe, 2-S ferredoxin from Pseudomonas.
Biochemistry, 33, 1994
|
|
1PUU
 
 | | Mistletoe lectin I in complex with lactose | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Krauspenhaar, R, Voelter, W, Stoeva, S, Mikhailov, A, Konareva, N, Betzel, C. | | Deposit date: | 2003-06-25 | | Release date: | 2004-06-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mistletoe lectin I in complex with galactose and lactose reveals distinct sugar-binding properties
Acta Crystallogr.,Sect.F, 61, 2005
|
|
1PUX
 
 | | NMR Solution Structure of BeF3-Activated Spo0F, 20 conformers | | Descriptor: | Sporulation initiation phosphotransferase F | | Authors: | Gardino, A.K, Volkman, B.F, Cho, H.S, Lee, S.Y, Wemmer, D.E, Kern, D. | | Deposit date: | 2003-06-25 | | Release date: | 2003-08-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The NMR solution structure of BeF(3)(-)-activated Spo0F reveals the conformational switch in a phosphorelay system.
J.Mol.Biol., 331, 2003
|
|
1PUY
 
 | | 1.5 A resolution structure of a synthetic DNA hairpin with a stilbenediether linker | | Descriptor: | 5'-D(*GP*TP*TP*TP*TP*GP*(S02)P*CP*AP*AP*AP*AP*C)-3', MAGNESIUM ION | | Authors: | Egli, M, Tereshko, V, Murshudov, G, Sanishvili, R, Liu, X, Lewis, F.D. | | Deposit date: | 2003-06-25 | | Release date: | 2003-10-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Face-to-face and edge-to-face pi-pi interactions in a synthetic DNA hairpin with a stilbenediether linker
J.Am.Chem.Soc., 125, 2003
|
|
1PUZ
 
 | | Solution NMR Structure of Protein NMA1147 from Neisseria meningitidis. Northeast Structural Genomics Consortium Target MR19 | | Descriptor: | conserved hypothetical protein | | Authors: | Liu, G, Xu, D, Sukumaran, D.K, Chiang, Y, Acton, T, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2003-06-25 | | Release date: | 2004-06-29 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the hypothetical protein NMA1147 from Neisseria meningitidis reveals a distinct 5-helix bundle.
Proteins, 55, 2004
|
|
1PV0
 
 | | Structure of the Sda antikinase | | Descriptor: | Sda | | Authors: | Rowland, S.L, Burkholder, W.F, Maciejewski, M.W, Grossman, A.D, King, G.F. | | Deposit date: | 2003-06-26 | | Release date: | 2004-04-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and mechanism of Sda: an inhibitor of the histidine kinases that regulate initiation of sporulation in Bacillus subtilis
Mol.Cell, 13, 2004
|
|
1PV1
 
 | | Crystal Structure Analysis of Yeast Hypothetical Protein: YJG8_YEAST | | Descriptor: | Hypothetical 33.9 kDa esterase in SMC3-MRPL8 intergenic region | | Authors: | Millard, C, Kumaran, D, Eswaramoorthy, S, Swaminathan, S, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2003-06-26 | | Release date: | 2004-11-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural characterization and reversal of the natural organophosphate resistance of a D-type esterase, Saccharomyces cerevisiae S-formylglutathione hydrolase.
Biochemistry, 47, 2008
|
|
1PV2
 
 | |
1PV3
 
 | | NMR Solution Structure of the Avian FAT-domain of Focal Adhesion Kinase | | Descriptor: | Focal adhesion kinase 1 | | Authors: | Prutzman, K.C, Gao, G, King, M.L, Iyer, V.V, Mueller, G.A, Schaller, M.D, Campbell, S.L. | | Deposit date: | 2003-06-26 | | Release date: | 2004-05-25 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The Focal Adhesion Targeting Domain of Focal Adhesion Kinase Contains a Hinge Region that Modulates Tyrosine 926 Phosphorylation.
STRUCTURE, 12, 2004
|
|
1PV4
 
 | |
1PV5
 
 | |
1PV6
 
 | | Crystal structure of lactose permease | | Descriptor: | Lactose permease | | Authors: | Abramson, J, Smirnova, I, Kasho, V, Verner, G, Kaback, H.R, Iwata, S. | | Deposit date: | 2003-06-26 | | Release date: | 2003-08-12 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure and mechanism of the lactose permease of Escherichia coli
SCIENCE, 301, 2003
|
|
1PV7
 
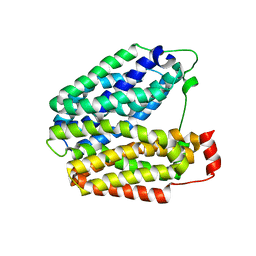 | | Crystal structure of lactose permease with TDG | | Descriptor: | Lactose permease, beta-D-galactopyranose-(1-1)-1-thio-beta-D-galactopyranose | | Authors: | Abramson, J, Smirnova, I, Kasho, V, Verner, G, Kaback, H.R, Iwata, S. | | Deposit date: | 2003-06-26 | | Release date: | 2003-08-12 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structure and mechanism of the lactose permease of Escherichia coli
SCIENCE, 301, 2003
|
|
1PV8
 
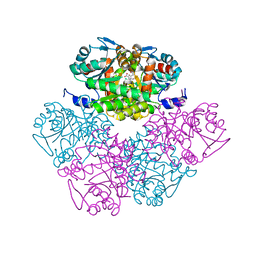 | | Crystal structure of a low activity F12L mutant of human porphobilinogen synthase | | Descriptor: | 3-(2-AMINOETHYL)-4-(AMINOMETHYL)HEPTANEDIOIC ACID, Delta-aminolevulinic acid dehydratase, ZINC ION | | Authors: | Breinig, S, Kervinen, J, Stith, L, Wasson, A.S, Fairman, R, Wlodawer, A, Zdanov, A, Jaffe, E.K. | | Deposit date: | 2003-06-26 | | Release date: | 2003-09-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Control of tetrapyrrole biosynthesis by alternate quaternary forms of porphobilinogen synthase.
Nat.Struct.Biol., 10, 2003
|
|
1PV9
 
 | | Prolidase from Pyrococcus furiosus | | Descriptor: | Xaa-Pro dipeptidase, ZINC ION | | Authors: | Maher, M.J, Ghosh, M, Grunden, A.M, Menon, A.L, Adams, M.W, Freeman, H.C, Guss, J.M. | | Deposit date: | 2003-06-27 | | Release date: | 2004-03-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the Prolidase from Pyrococcus furiosus.
Biochemistry, 43, 2004
|
|
1PVA
 
 | | COMPARISON BETWEEN THE CRYSTAL AND THE SOLUTION STRUCTURES OF THE EF HAND PARVALBUMIN (ALPHA COMPONENT FROM PIKE MUSCLE) | | Descriptor: | CALCIUM ION, PARVALBUMIN | | Authors: | Declercq, J.P, Tinant, B, Roquet, F, Rambaud, J, Parello, J. | | Deposit date: | 1995-01-16 | | Release date: | 1995-03-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Comparison between the Crystal and the Solution Structures of the EF Hand Parvalbumin (Alpha Component from Pike Muscle)
To be Published
|
|
1PVB
 
 | | X-RAY STRUCTURE OF A NEW CRYSTAL FORM OF PIKE 4.10 PARVALBUMIN | | Descriptor: | AMMONIUM ION, CALCIUM ION, PARVALBUMIN | | Authors: | Declercq, J.P, Tinant, B, Parello, J. | | Deposit date: | 1995-01-05 | | Release date: | 1995-02-27 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | X-ray structure of a new crystal form of pike 4.10 beta parvalbumin.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
1PVC
 
 | |
1PVD
 
 | |
1PVE
 
 | |
1PVF
 
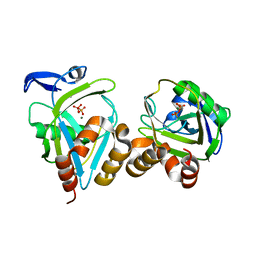 | | E.coli IPP isomerase in complex with diphosphate | | Descriptor: | DIPHOSPHATE, Isopentenyl-diphosphate delta-isomerase, MAGNESIUM ION, ... | | Authors: | Wouters, J, Durisotti, V, Oudjama, Y, Stalon, V, Droogmans, L. | | Deposit date: | 2003-06-27 | | Release date: | 2004-07-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | E.coli Isopentenyl Diphosphate: Dimethylallyl Diphosphate isomerase in complex with diphosphate
To be Published
|
|
1PVG
 
 | |
