1L9Z
 
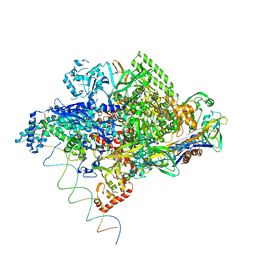 | | Thermus aquaticus RNA Polymerase Holoenzyme/Fork-Junction Promoter DNA Complex at 6.5 A Resolution | | Descriptor: | MAGNESIUM ION, RNA POLYMERASE, ALPHA SUBUNIT, ... | | Authors: | Murakami, K.S, Masuda, S, Campbell, E.A, Muzzin, O, Darst, S.A. | | Deposit date: | 2002-03-27 | | Release date: | 2002-05-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (6.5 Å) | | Cite: | Structural basis of transcription initiation: an RNA polymerase holoenzyme-DNA complex.
Science, 296, 2002
|
|
1LA0
 
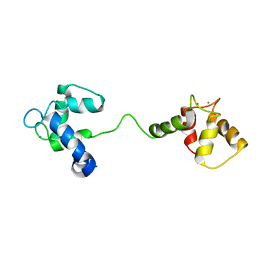 | | Solution Structure of Calcium Saturated Cardiac Troponin C in the Troponin C-Troponin I Complex | | Descriptor: | CALCIUM ION, Troponin C, slow skeletal and cardiac muscles | | Authors: | Dvoretsky, A, Abusamhadneh, E.M, Howarth, J.W, Rosevear, P.R. | | Deposit date: | 2002-03-27 | | Release date: | 2002-12-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Calcium-Saturated Cardiac Troponin C bound to cardiac Troponin I.
J.Biol.Chem., 277, 2002
|
|
1LA1
 
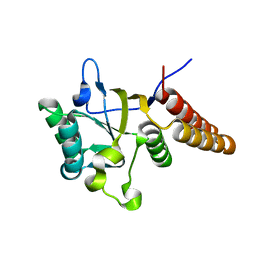 | | Gro-EL Fragment (Apical Domain) Comprising Residues 188-379 | | Descriptor: | GroEL | | Authors: | Ashcroft, A.E, Brinker, A, Coyle, J.E, Weber, F, Kaiser, M, Moroder, L, Parsons, M.R, Jager, J, Hartl, U.F, Hayer-Hartl, M, Radford, S.E. | | Deposit date: | 2002-03-27 | | Release date: | 2002-04-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structural plasticity and noncovalent substrate binding in the GroEL apical domain. A study using electrospay ionization mass spectrometry and fluorescence binding studies.
J.Biol.Chem., 277, 2002
|
|
1LA2
 
 | | Structural analysis of Saccharomyces cerevisiae myo-inositol phosphate synthase | | Descriptor: | Myo-inositol-1-phosphate synthase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Kniewel, R, Buglino, J.A, Shen, V, Chadna, T, Beckwith, A, Lima, C.D, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2002-03-27 | | Release date: | 2002-04-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural analysis of Saccharomyces cerevisiae myo-inositol phosphate synthase
J.STRUCT.FUNCT.GENOM., 2, 2002
|
|
1LA3
 
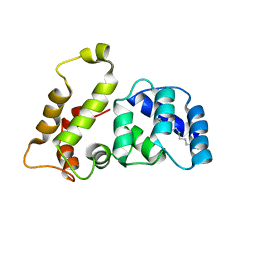 | | Solution structure of recoverin mutant, E85Q | | Descriptor: | CALCIUM ION, MYRISTIC ACID, Recoverin | | Authors: | Ames, J.B, Hamasaki, N, Molchanova, T. | | Deposit date: | 2002-03-27 | | Release date: | 2002-06-19 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Structure and calcium-binding studies of a recoverin mutant (E85Q) in an allosteric intermediate state.
Biochemistry, 41, 2002
|
|
1LA4
 
 | | Solution Structure of SGTx1 | | Descriptor: | SGTx1 | | Authors: | Lee, C.W, Roh, S.H, Kim, S, Endoh, H, Kodera, Y, Maeda, T, Swartz, K.J, Kim, J.I. | | Deposit date: | 2002-03-28 | | Release date: | 2003-11-11 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Solution Structure and Functional Characterization of SGTx1, a Modifier of Kv2.1 Channel Gating
Biochemistry, 43, 2004
|
|
1LA6
 
 | | The crystal structure of Trematomus newnesi hemoglobin in a partial hemichrome state | | Descriptor: | CARBON MONOXIDE, Hemoglobin alpha-1 chain, Hemoglobin beta-1/2 chain, ... | | Authors: | Riccio, A, Vitagliano, L, di Prisco, G, Zagari, A, Mazzarella, L. | | Deposit date: | 2002-03-28 | | Release date: | 2002-07-31 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of a tetrameric hemoglobin in a partial hemichrome state
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1LA8
 
 | | Solution structure of the DNA hairpin 13-mer CGCGGTGTCCGCG | | Descriptor: | 5'-D(*CP*GP*CP*GP*GP*TP*GP*TP*CP*CP*GP*CP*G)-3' | | Authors: | Weisenseel, J.P, Reddy, G.R, Marnett, L.J, Stone, M.P. | | Deposit date: | 2002-03-28 | | Release date: | 2002-04-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the 1,N(2)-propanodeoxyguanosine adduct in a three-base DNA hairpin loop derived from a palindrome in the Salmonella typhimurium hisD3052 gene.
Chem.Res.Toxicol., 15, 2002
|
|
1LAA
 
 | |
1LAB
 
 | |
1LAC
 
 | |
1LAE
 
 | | Solution Structure of the DNA 13-mer Hairpin CGCGGTXTCCGCG (X=PdG) Containing the 1,N2-propanodeoxyguanosine Adduct at the Seventh Position | | Descriptor: | 5'-D(*CP*GP*CP*GP*GP*TP*(P)P*TP*CP*CP*GP*CP*G)-3' | | Authors: | Weisenseel, J.P, Reddy, G.R, Marnett, L.J, Stone, M.P. | | Deposit date: | 2002-03-28 | | Release date: | 2002-04-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the 1,N(2)-propanodeoxyguanosine adduct in a three-base DNA hairpin loop derived from a palindrome in the Salmonella typhimurium hisD3052 gene.
Chem.Res.Toxicol., 15, 2002
|
|
1LAF
 
 | | STRUCTURAL BASES FOR MULTIPLE LIGAND SPECIFICITY OF THE PERIPLASMIC LYSINE-, ARGININE-, ORNITHINE-BINDING PROTEIN | | Descriptor: | ARGININE, LYSINE, ORNITHINE-BINDING PROTEIN | | Authors: | Kim, S.-H, Oh, B.-H. | | Deposit date: | 1993-10-06 | | Release date: | 1995-07-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structural basis for multiple ligand specificity of the periplasmic lysine-, arginine-, ornithine-binding protein.
J.Biol.Chem., 269, 1994
|
|
1LAG
 
 | | STRUCTURAL BASES FOR MULTIPLE LIGAND SPECIFICITY OF THE PERIPLASMIC LYSINE-, ARGININE-, ORNITHINE-BINDING PROTEIN | | Descriptor: | HISTIDINE, LYSINE, ARGININE, ... | | Authors: | Kim, S.-H, Oh, B.-H. | | Deposit date: | 1993-10-06 | | Release date: | 1995-07-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structural basis for multiple ligand specificity of the periplasmic lysine-, arginine-, ornithine-binding protein.
J.Biol.Chem., 269, 1994
|
|
1LAH
 
 | | STRUCTURAL BASES FOR MULTIPLE LIGAND SPECIFICITY OF THE PERIPLASMIC LYSINE-, ARGININE-, ORNITHINE-BINDING PROTEIN | | Descriptor: | L-ornithine, LYSINE, ARGININE, ... | | Authors: | Kim, S.-H, Oh, B.-H. | | Deposit date: | 1993-10-06 | | Release date: | 1995-07-10 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structural basis for multiple ligand specificity of the periplasmic lysine-, arginine-, ornithine-binding protein.
J.Biol.Chem., 269, 1994
|
|
1LAI
 
 | | Solution Structure of the B-DNA Duplex CGCGGTGTCCGCG. | | Descriptor: | 5'-D(*CP*GP*CP*GP*GP*AP*CP*AP*CP*CP*GP*CP*G)-3', 5'-D(*CP*GP*CP*GP*GP*TP*GP*TP*CP*CP*GP*CP*G)-3' | | Authors: | Weisenseel, J.P, Reddy, G.R, Marnett, L.J, Stone, M.P. | | Deposit date: | 2002-03-28 | | Release date: | 2002-04-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of an oligodeoxynucleotide containing a 1,N(2)-propanodeoxyguanosine adduct positioned in a palindrome derived from the Salmonella typhimurium hisD3052 gene: Hoogsteen pairing at pH 5.2.
Chem.Res.Toxicol., 15, 2002
|
|
1LAJ
 
 | | The Structure of Tomato Aspermy Virus by X-Ray Crystallography | | Descriptor: | 5'-R(*AP*AP*A)-3', MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Lucas, R.W, Larson, S.B, Canady, M.A, McPherson, A. | | Deposit date: | 2002-03-28 | | Release date: | 2002-11-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | The Structure of Tomato Aspermy Virus by X-Ray Crystallography
J.STRUCT.BIOL., 139, 2002
|
|
1LAM
 
 | | LEUCINE AMINOPEPTIDASE (UNLIGATED) | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, CARBONATE ION, LEUCINE AMINOPEPTIDASE, ... | | Authors: | Straeter, N, Lipscomb, W.N. | | Deposit date: | 1995-08-11 | | Release date: | 1995-10-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Two-metal ion mechanism of bovine lens leucine aminopeptidase: active site solvent structure and binding mode of L-leucinal, a gem-diolate transition state analogue, by X-ray crystallography.
Biochemistry, 34, 1995
|
|
1LAN
 
 | | LEUCINE AMINOPEPTIDASE COMPLEX WITH L-LEUCINAL | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, LEUCINE, LEUCINE AMINOPEPTIDASE, ... | | Authors: | Straeter, N, Lipscomb, W.N. | | Deposit date: | 1995-08-11 | | Release date: | 1995-10-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Two-metal ion mechanism of bovine lens leucine aminopeptidase: active site solvent structure and binding mode of L-leucinal, a gem-diolate transition state analogue, by X-ray crystallography.
Biochemistry, 34, 1995
|
|
1LAP
 
 | | MOLECULAR STRUCTURE OF LEUCINE AMINOPEPTIDASE AT 2.7-ANGSTROMS RESOLUTION | | Descriptor: | Cytosol aminopeptidase, ZINC ION | | Authors: | Burley, S.K, David, P.R, Taylor, A, Lipscomb, W.N. | | Deposit date: | 1990-08-01 | | Release date: | 1991-10-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular structure of leucine aminopeptidase at 2.7-A resolution.
Proc.Natl.Acad.Sci.USA, 87, 1990
|
|
1LAQ
 
 | | Solution Structure of the B-DNA Duplex CGCGGTXTCCGCG (X=PdG) Containing the 1,N2-propanodeoxyguanosine Adduct with the Deoxyribose at C20 Opposite PdG in the C2' Endo Conformation. | | Descriptor: | 5'-D(*CP*GP*CP*GP*GP*AP*(DNR)P*AP*CP*CP*GP*CP*G)-3', 5'-D(*CP*GP*CP*GP*GP*TP*(P)P*TP*CP*CP*GP*CP*G)-3' | | Authors: | Weisenseel, J.P, Reddy, G.R, Marnett, L.J, Stone, M.P. | | Deposit date: | 2002-03-29 | | Release date: | 2002-04-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of an oligodeoxynucleotide containing a 1,N(2)-propanodeoxyguanosine adduct positioned in a palindrome derived from the Salmonella typhimurium hisD3052 gene: Hoogsteen pairing at pH 5.2.
Chem.Res.Toxicol., 15, 2002
|
|
1LAR
 
 | | CRYSTAL STRUCTURE OF THE TANDEM PHOSPHATASE DOMAINS OF RPTP LAR | | Descriptor: | PROTEIN (LAR) | | Authors: | Nam, H.-J, Poy, F, Krueger, N, Saito, H, Frederick, C.A. | | Deposit date: | 1999-04-20 | | Release date: | 2000-04-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the tandem phosphatase domains of RPTP LAR.
Cell(Cambridge,Mass.), 97, 1999
|
|
1LAS
 
 | | Solution Structure of the B-DNA Duplex CGCGGTXTCCGCG (X=PdG) Containing the 1,N2-propanodeoxyguanosine Adduct with the Deoxyribose at C20 Opposite PdG in the C3' Endo Conformation. | | Descriptor: | 5'-D(*CP*GP*CP*GP*GP*AP*(DNR)P*AP*CP*CP*GP*CP*G)-3', 5'-D(*CP*GP*CP*GP*GP*TP*(P)P*TP*CP*CP*GP*CP*G)-3' | | Authors: | Weisenseel, J.P, Reddy, G.R, Marnett, L.J, Stone, M.P. | | Deposit date: | 2002-03-29 | | Release date: | 2002-04-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of an oligodeoxynucleotide containing a 1,N(2)-propanodeoxyguanosine adduct positioned in a palindrome derived from the Salmonella typhimurium hisD3052 gene: Hoogsteen pairing at pH 5.2.
Chem.Res.Toxicol., 15, 2002
|
|
1LAT
 
 | | GLUCOCORTICOID RECEPTOR MUTANT/DNA COMPLEX | | Descriptor: | DNA (5'-D(*TP*TP*CP*CP*AP*GP*AP*AP*CP*AP*TP*GP*TP*TP*CP*TP*G P*GP*A)-3'), GLUCOCORTICOID RECEPTOR, ZINC ION | | Authors: | Gewirth, D.T, Sigler, P.B. | | Deposit date: | 1995-12-18 | | Release date: | 1996-04-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The basis for half-site specificity explored through a non-cognate steroid receptor-DNA complex.
Nat.Struct.Biol., 2, 1995
|
|
1LAU
 
 | | URACIL-DNA GLYCOSYLASE | | Descriptor: | DNA (5'-D(*TP*TP*T)-3'), PROTEIN (URACIL-DNA GLYCOSYLASE (E.C.3.2.2.-)) | | Authors: | Pearl, L.H, Savva, R. | | Deposit date: | 1996-01-03 | | Release date: | 1996-06-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structural basis of specific base-excision repair by uracil-DNA glycosylase.
Nature, 373, 1995
|
|
