1L62
 
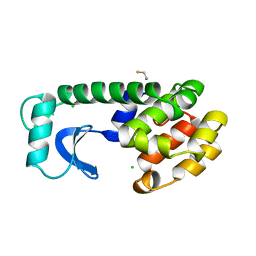 | |
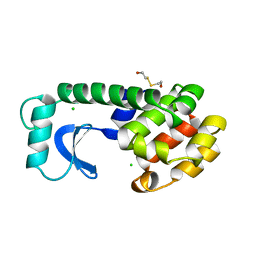
1L63
 
 | |
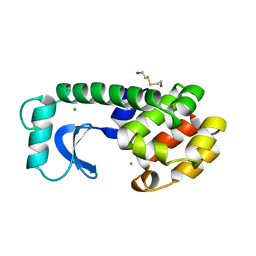
1L64
 
 | |
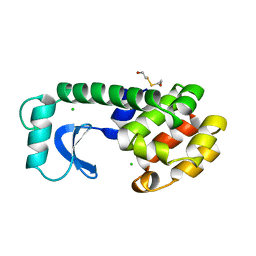
1L65
 
 | |
1L66
 
 | |
1L67
 
 | |
1L68
 
 | |
1L69
 
 | |
1L6B
 
 | |
1L6E
 
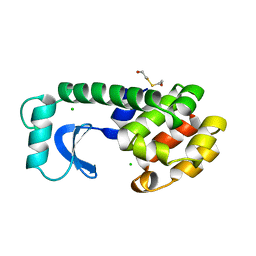
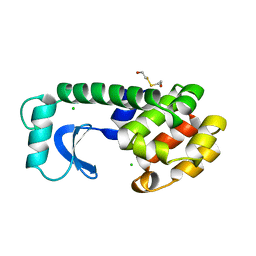
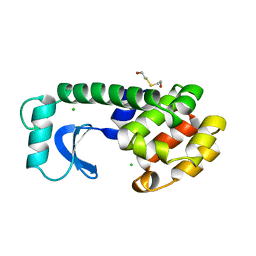
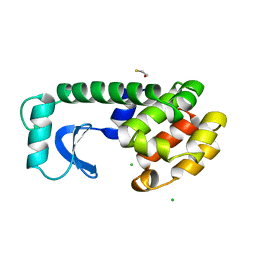
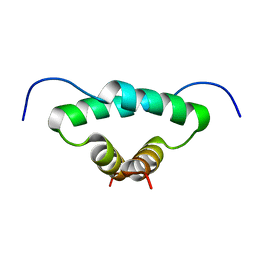 | | Solution structure of the docking and dimerization domain of protein kinase A II-alpha (RIIalpha D/D). Alternatively called the N-terminal dimerization domain of the regulatory subunit of protein kinase A. | | Descriptor: | cAMP-dependent protein kinase Type II-alpha regulatory chain | | Authors: | Morikis, D, Roy, M, Newlon, M.G, Scott, J.D, Jennings, P.A. | | Deposit date: | 2002-03-08 | | Release date: | 2002-04-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Electrostatic properties of the structure of the docking and dimerization domain of protein kinase A IIalpha
Eur.J.Biochem., 269, 2002
|
|
1L6F
 
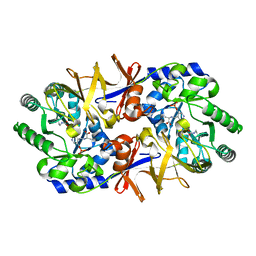 | | Alanine racemase bound with N-(5'-phosphopyridoxyl)-L-alanine | | Descriptor: | ALANYL-PYRIDOXAL-5'-PHOSPHATE, alanine racemase | | Authors: | Watanabe, A, Yoshimura, T, Mikami, B, Hayashi, H, Kagamiyama, H, Esaki, N. | | Deposit date: | 2002-03-09 | | Release date: | 2002-06-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Reaction mechanism of alanine racemase from Bacillus stearothermophilus: x-ray crystallographic studies of the enzyme bound with N-(5'-phosphopyridoxyl)alanine.
J.Biol.Chem., 277, 2002
|
|
1L6G
 
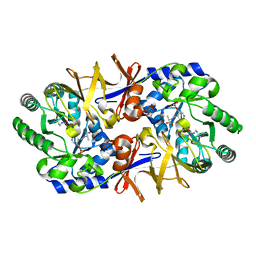 | | Alanine racemase bound with N-(5'-phosphopyridoxyl)-D-alanine | | Descriptor: | N-(5'-PHOSPHOPYRIDOXYL)-D-ALANINE, alanine racemase | | Authors: | Watanabe, A, Yoshimura, T, Mikami, B, Hayashi, H, Kagamiyama, H, Esaki, N. | | Deposit date: | 2002-03-10 | | Release date: | 2002-06-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Reaction mechanism of alanine racemase from Bacillus stearothermophilus: x-ray crystallographic studies of the enzyme bound with N-(5'-phosphopyridoxyl)alanine.
J.Biol.Chem., 277, 2002
|
|
1L6H
 
 | |
1L6I
 
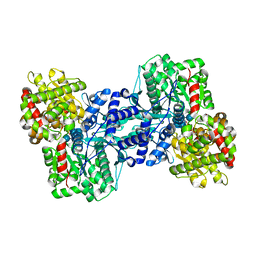 | | Crystal Structure of the Maltodextrin Phosphorylase complexed with the products of the enzymatic reaction between glucose-1-phosphate and maltopentaose | | Descriptor: | MALTODEXTRIN PHOSPHORYLASE, PHOSPHATE ION, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Geremia, S, Campagnolo, M, Schinzel, R, Johnson, L.N. | | Deposit date: | 2002-03-11 | | Release date: | 2002-04-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Enzymatic catalysis in crystals of Escherichia coli maltodextrin phosphorylase
J.Mol.Biol., 322, 2002
|
|
1L6J
 
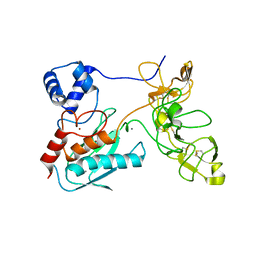 | | Crystal structure of human matrix metalloproteinase MMP9 (gelatinase B). | | Descriptor: | CALCIUM ION, Matrix metalloproteinase-9, ZINC ION | | Authors: | Elkins, P.A, Ho, Y.S, Smith, W.W, Janson, C.A, D'Alessio, K.J, McQueney, M.S, Cummings, M.D, Romanic, A.M. | | Deposit date: | 2002-03-11 | | Release date: | 2002-07-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the C-terminally truncated human ProMMP9, a gelatin-binding matrix metalloproteinase.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1L6M
 
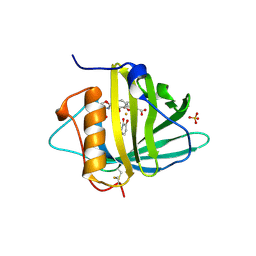 | | Neutrophil Gelatinase-associated Lipocalin is a Novel Bacteriostatic Agent that Interferes with Siderophore-mediated Iron Acquisition | | Descriptor: | 2,3-DIHYDROXY-BENZOIC ACID, 2-(2,3-DIHYDROXY-BENZOYLAMINO)-3-HYDROXY-PROPIONIC ACID, FE (III) ION, ... | | Authors: | Goetz, D.H, Borregaard, N, Bluhm, M.E, Raymond, K.N, Strong, R.K. | | Deposit date: | 2002-03-11 | | Release date: | 2003-03-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Neutrophil Lipocalin NGAL is a Bacteriostatic Agent that Interferes with Siderophore-mediated Iron Acquisition
Mol.Cell, 10, 2002
|
|
1L6N
 
 | |
1L6O
 
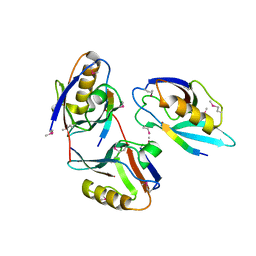 | | XENOPUS DISHEVELLED PDZ DOMAIN | | Descriptor: | Dapper 1, Segment polarity protein dishevelled homolog DVL-2 | | Authors: | Cheyette, B.N.R, Waxman, J.S, Miller, J.R, Takemaru, K.-I, Sheldahl, L.C, Khlebtsova, N, Fox, E.P, Earnest, T, Moon, R.T. | | Deposit date: | 2002-03-11 | | Release date: | 2003-06-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Dapper, a Dishevelled-associated antagonist of beta-catenin and JNK signaling, is required for notochord formation
Dev.Cell, 2, 2002
|
|
1L6P
 
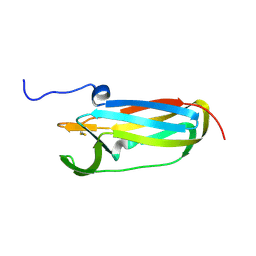 | |
1L6R
 
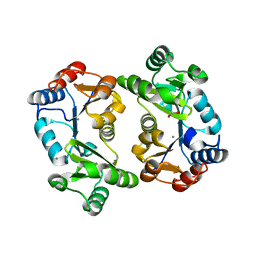 | | Crystal Structure of Thermoplasma acidophilum 0175 (APC0014) | | Descriptor: | CALCIUM ION, FORMIC ACID, HYPOTHETICAL PROTEIN TA0175 | | Authors: | Kim, Y, Joachimiak, A, Edwards, A.M, Xu, X, Pennycooke, M, Gu, J, Cheung, F, Christendat, D, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-03-13 | | Release date: | 2003-01-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure- and function-based characterization of a new phosphoglycolate phosphatase from Thermoplasma acidophilum.
J.Biol.Chem., 279, 2004
|
|
1L6S
 
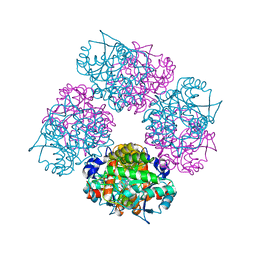 | | Crystal Structure of Porphobilinogen Synthase Complexed with the Inhibitor 4,7-Dioxosebacic Acid | | Descriptor: | 4,7-DIOXOSEBACIC ACID, MAGNESIUM ION, PORPHOBILINOGEN SYNTHASE, ... | | Authors: | Jaffe, E.K, Kervinen, J, Martins, J, Stauffer, F, Neier, R, Wlodawer, A, Zdanov, A. | | Deposit date: | 2002-03-13 | | Release date: | 2002-04-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Species-Specific Inhibition of Porphobilinogen Synthase by 4-Oxosebacic Acid
J.Biol.Chem., 277, 2002
|
|
1L6T
 
 | |
1L6U
 
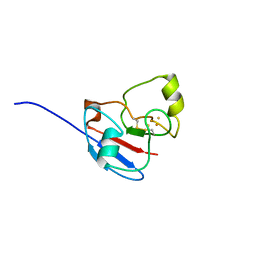 | | NMR STRUCTURE OF OXIDIZED ADRENODOXIN | | Descriptor: | Adrenodoxin 1, FE2/S2 (INORGANIC) CLUSTER | | Authors: | Beilke, D, Weiss, R, Lohr, F, Pristovsek, P, Hannemann, F, Bernhardt, R, Rueterjans, H. | | Deposit date: | 2002-03-14 | | Release date: | 2002-06-26 | | Last modified: | 2020-02-05 | | Method: | SOLUTION NMR | | Cite: | A new electron transport mechanism in mitochondrial steroid hydroxylase systems based on structural changes upon the reduction of adrenodoxin.
Biochemistry, 41, 2002
|
|
1L6V
 
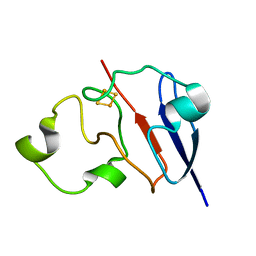 | | STRUCTURE OF REDUCED BOVINE ADRENODOXIN | | Descriptor: | Adrenodoxin 1, FE2/S2 (INORGANIC) CLUSTER | | Authors: | Beilke, D, Weiss, R, Lohr, F, Pristovsek, P, Hannemann, F, Bernhardt, R, Rueterjans, H. | | Deposit date: | 2002-03-14 | | Release date: | 2002-06-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A new electron transport mechanism in mitochondrial steroid hydroxylase systems based on structural changes upon the reduction of adrenodoxin.
Biochemistry, 41, 2002
|
|
1L6W
 
 | | Fructose-6-phosphate aldolase | | Descriptor: | Fructose-6-phosphate aldolase 1, GLYCEROL | | Authors: | Thorell, S, Schuermann, M, Sprenger, G.A, Schneider, G. | | Deposit date: | 2002-03-14 | | Release date: | 2002-06-12 | | Last modified: | 2018-03-07 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structure of decameric fructose-6-phosphate aldolase from Escherichia coli reveals inter-subunit helix swapping as a structural basis for assembly differences in the transaldolase family.
J.Mol.Biol., 319, 2002
|
|
