1AJC
 
 | |
1AJD
 
 | |
1AJE
 
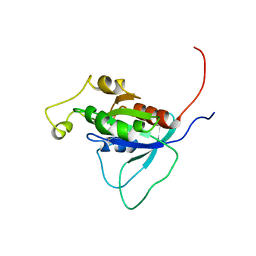 | | CDC42 FROM HUMAN, NMR, 20 STRUCTURES | | Descriptor: | CDC42HS | | Authors: | Feltham, J.L, Dotsch, V, Raza, S, Manor, D, Cerione, R.A, Sutcliffe, M.J, Wagner, G, Oswald, R.E. | | Deposit date: | 1997-05-02 | | Release date: | 1997-11-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Definition of the switch surface in the solution structure of Cdc42Hs.
Biochemistry, 36, 1997
|
|
1AJF
 
 | |
1AJG
 
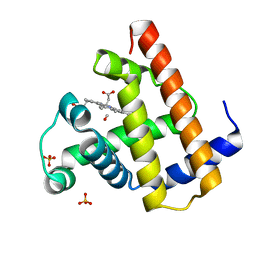 | | CARBONMONOXY MYOGLOBIN AT 40 K | | Descriptor: | CARBON MONOXIDE, MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Teng, T.Y, Srajer, V, Moffat, K. | | Deposit date: | 1997-05-02 | | Release date: | 1997-11-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Photolysis-induced structural changes in single crystals of carbonmonoxy myoglobin at 40 K.
Nat.Struct.Biol., 1, 1994
|
|
1AJH
 
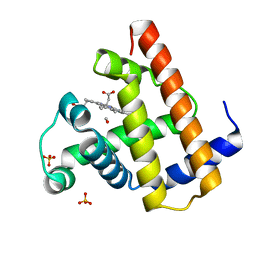 | | PHOTOPRODUCT OF CARBONMONOXY MYOGLOBIN AT 40 K | | Descriptor: | CARBON MONOXIDE, MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Teng, T.Y, Srajer, V, Moffat, K. | | Deposit date: | 1997-05-02 | | Release date: | 1997-11-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Photolysis-induced structural changes in single crystals of carbonmonoxy myoglobin at 40 K.
Nat.Struct.Biol., 1, 1994
|
|
1AJJ
 
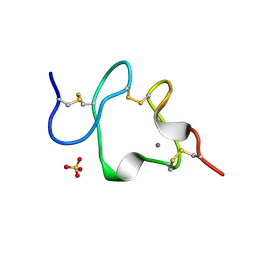 | | LDL RECEPTOR LIGAND-BINDING MODULE 5, CALCIUM-COORDINATING | | Descriptor: | CALCIUM ION, LOW-DENSITY LIPOPROTEIN RECEPTOR, SULFATE ION | | Authors: | Fass, D, Blacklow, S.C, Kim, P.S, Berger, J.M. | | Deposit date: | 1997-05-04 | | Release date: | 1997-07-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular basis of familial hypercholesterolaemia from structure of LDL receptor module.
Nature, 388, 1997
|
|
1AJK
 
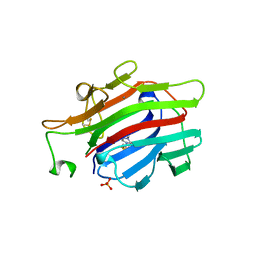 | | CIRCULARLY PERMUTED (1-3,1-4)-BETA-D-GLUCAN 4-GLUCANOHYDROLASE CPA16M-84 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, CIRCULARLY PERMUTED (1-3,1-4)-BETA-D-GLUCAN 4-GLUCANOHYDROLASE, ... | | Authors: | Ay, J, Heinemann, U. | | Deposit date: | 1997-05-06 | | Release date: | 1998-05-06 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures and properties of de novo circularly permuted 1,3-1,4-beta-glucanases.
Proteins, 30, 1998
|
|
1AJL
 
 | |
1AJM
 
 | |
1AJN
 
 | | PENICILLIN ACYLASE COMPLEXED WITH P-NITROPHENYLACETIC ACID | | Descriptor: | 2-(4-NITROPHENYL)ACETIC ACID, CALCIUM ION, PENICILLIN AMIDOHYDROLASE | | Authors: | Done, S.H. | | Deposit date: | 1997-05-07 | | Release date: | 1997-11-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Ligand-induced conformational change in penicillin acylase.
J.Mol.Biol., 284, 1998
|
|
1AJO
 
 | | CIRCULARLY PERMUTED (1-3,1-4)-BETA-D-GLUCAN 4-GLUCANOHYDROLASE CPA16M-127 | | Descriptor: | CALCIUM ION, CIRCULARLY PERMUTED (1-3,1-4)-BETA-D-GLUCAN 4-GLUCANOHYDROLASE CPA16M-127 | | Authors: | Ay, J, Heinemann, U. | | Deposit date: | 1997-05-07 | | Release date: | 1998-05-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Crystal structures and properties of de novo circularly permuted 1,3-1,4-beta-glucanases.
Proteins, 30, 1998
|
|
1AJP
 
 | |
1AJQ
 
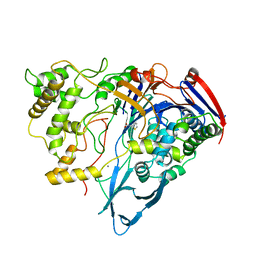 | | PENICILLIN ACYLASE COMPLEXED WITH THIOPHENEACETIC ACID | | Descriptor: | CALCIUM ION, PENICILLIN AMIDOHYDROLASE, THIOPHENEACETIC ACID | | Authors: | Done, S.H. | | Deposit date: | 1997-05-07 | | Release date: | 1997-11-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Ligand-induced conformational change in penicillin acylase.
J.Mol.Biol., 284, 1998
|
|
1AJR
 
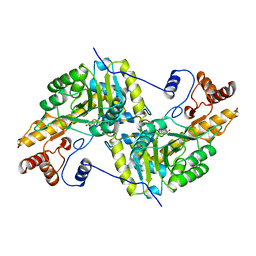 | | REFINEMENT AND COMPARISON OF THE CRYSTAL STRUCTURES OF PIG CYTOSOLIC ASPARTATE AMINOTRANSFERASE AND ITS COMPLEX WITH 2-METHYLASPARTATE | | Descriptor: | ASPARTATE AMINOTRANSFERASE | | Authors: | Rhee, S, Silva, M.M, Hyde, C.C, Rogers, P.H, Metzler, C.M, Metzler, D.E, Arnone, A. | | Deposit date: | 1997-05-08 | | Release date: | 1997-08-20 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Refinement and comparisons of the crystal structures of pig cytosolic aspartate aminotransferase and its complex with 2-methylaspartate.
J.Biol.Chem., 272, 1997
|
|
1AJS
 
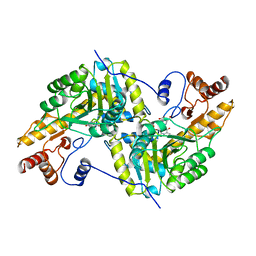 | | REFINEMENT AND COMPARISON OF THE CRYSTAL STRUCTURES OF PIG CYTOSOLIC ASPARTATE AMINOTRANSFERASE AND ITS COMPLEX WITH 2-METHYLASPARTATE | | Descriptor: | 2-[(3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YLMETHYL)-AMINO]-2-METHYL-SUCCINIC ACID, ASPARTATE AMINOTRANSFERASE | | Authors: | Rhee, S, Silva, M.M, Hyde, C.C, Rogers, P.H, Metzler, C.M, Metzler, D.E, Arnone, A. | | Deposit date: | 1997-05-08 | | Release date: | 1997-08-20 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Refinement and comparisons of the crystal structures of pig cytosolic aspartate aminotransferase and its complex with 2-methylaspartate.
J.Biol.Chem., 272, 1997
|
|
1AJT
 
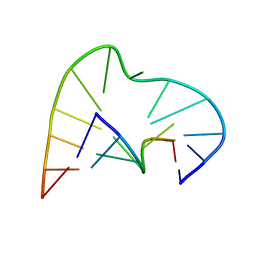 | | FIVE-NUCLEOTIDE BULGE LOOP FROM TETRAHYMENA THERMOPHILA GROUP I INTRON, NMR, 1 STRUCTURE | | Descriptor: | RNA (5'-R(*GP*AP*GP*UP*AP*CP*C)-3'), RNA (5'-R(*GP*GP*UP*AP*AP*UP*AP*AP*GP*CP*UP*C)-3') | | Authors: | Luebke, K.J, Landry, S.M, Tinoco Junior, I. | | Deposit date: | 1997-05-08 | | Release date: | 1997-08-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution conformation of a five-nucleotide RNA bulge loop from a group I intron.
Biochemistry, 36, 1997
|
|
1AJU
 
 | |
1AJV
 
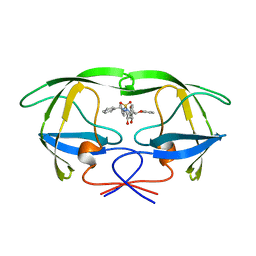 | | HIV-1 PROTEASE IN COMPLEX WITH THE CYCLIC SULFAMIDE INHIBITOR AHA006 | | Descriptor: | 2,7-DIBENZYL-1,1-DIOXO-3,6-BIS-PHENOXYMETHYL-[1,2,7]THIADIAZEPANE-4,5-DIOL, HIV-1 PROTEASE | | Authors: | Backbro, K, Unge, T. | | Deposit date: | 1997-05-11 | | Release date: | 1997-08-20 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Unexpected binding mode of a cyclic sulfamide HIV-1 protease inhibitor.
J.Med.Chem., 40, 1997
|
|
1AJW
 
 | |
1AJX
 
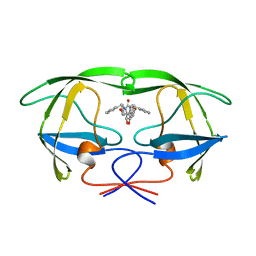 | |
1AJY
 
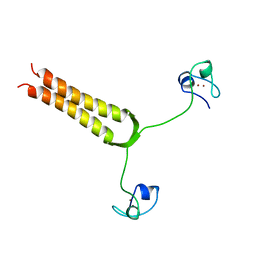 | | STRUCTURE AND MOBILITY OF THE PUT3 DIMER: A DNA PINCER, NMR, 13 STRUCTURES | | Descriptor: | PUT3, ZINC ION | | Authors: | Walters, K.J, Dayie, K.T, Reece, R.J, Ptashne, M, Wagner, G. | | Deposit date: | 1997-05-12 | | Release date: | 1997-09-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and mobility of the PUT3 dimer.
Nat.Struct.Biol., 4, 1997
|
|
1AJZ
 
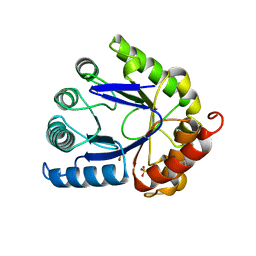 | | STRUCTURE OF DIHYDROPTEROATE PYROPHOSPHORYLASE | | Descriptor: | DIHYDROPTEROATE SYNTHASE, SULFATE ION | | Authors: | Achari, A, Somers, D.O, Champness, J.N, Bryant, P.K, Rosemond, J, Stammers, D.K. | | Deposit date: | 1997-05-13 | | Release date: | 1998-05-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the anti-bacterial sulfonamide drug target dihydropteroate synthase.
Nat.Struct.Biol., 4, 1997
|
|
1AK0
 
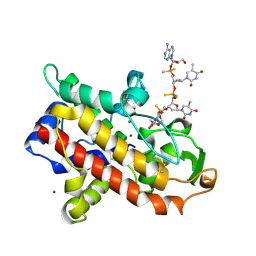 | | P1 NUCLEASE IN COMPLEX WITH A SUBSTRATE ANALOG | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-(DITHIO)PHOSPHATE, ... | | Authors: | Romier, C, Suck, D. | | Deposit date: | 1997-05-28 | | Release date: | 1997-12-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Recognition of single-stranded DNA by nuclease P1: high resolution crystal structures of complexes with substrate analogs.
Proteins, 32, 1998
|
|
1AK1
 
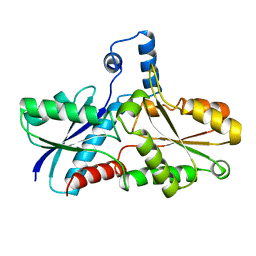 | | FERROCHELATASE FROM BACILLUS SUBTILIS | | Descriptor: | FERROCHELATASE | | Authors: | Al-Karadaghi, S, Hansson, M, Nikonov, S, Jonsson, B, Hederstedt, L. | | Deposit date: | 1997-05-28 | | Release date: | 1997-12-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of ferrochelatase: the terminal enzyme in heme biosynthesis.
Structure, 5, 1997
|
|
