6KLK
 
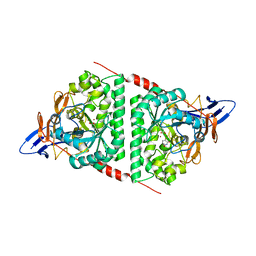 | |
5VGM
 
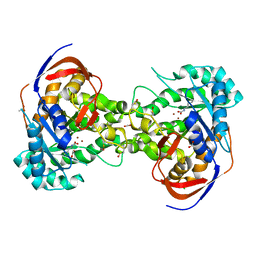 | | Crystal structure of dihydroorotase pyrC from Vibrio cholerae in complex with zinc at 1.95 A resolution. | | Descriptor: | ACETATE ION, CHLORIDE ION, Dihydroorotase, ... | | Authors: | Lipowska, J, Shabalin, I.G, Miks, C.D, Winsor, J, Cooper, D.R, Shuvalova, L, Kwon, K, Lewinski, K, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-04-11 | | Release date: | 2017-04-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Pyrimidine biosynthesis in pathogens - Structures and analysis of dihydroorotases from Yersinia pestis and Vibrio cholerae.
Int.J.Biol.Macromol., 136, 2019
|
|
6CTY
 
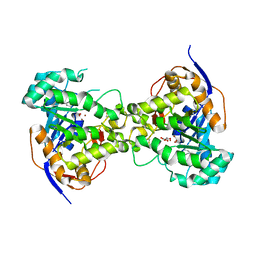 | | Crystal structure of dihydroorotase pyrC from Yersinia pestis in complex with zinc and malate at 2.4 A resolution | | Descriptor: | D-MALATE, Dihydroorotase, ZINC ION | | Authors: | Lipowska, J, Shabalin, I.G, Winsor, J, Woinska, M, Cooper, D.R, Kwon, K, Shuvalova, L, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-03-23 | | Release date: | 2018-04-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Pyrimidine biosynthesis in pathogens - Structures and analysis of dihydroorotases from Yersinia pestis and Vibrio cholerae.
Int.J.Biol.Macromol., 136, 2019
|
|
7X68
 
 | | CYS179 and CYS504 of CRMP2 were covalently binded by a Sesquiterpene lactone | | Descriptor: | (3aR,5S,8R,8aR,9aR)-5,8a-dimethyl-3-methylidene-8-oxidanyl-5,6,7,8,9,9a-hexahydro-3aH-benzo[f][1]benzofuran-2-one, Dihydropyrimidinase-related protein 2, SODIUM ION | | Authors: | Zhang, S.D, Ma, Y.F, Zhang, J. | | Deposit date: | 2022-03-06 | | Release date: | 2022-05-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | CYS179 and CYS504 of CRMP2 were covalently binded by a Sesquiterpene lactone
To Be Published
|
|
6AJD
 
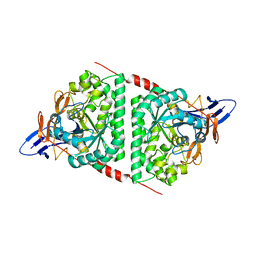 | |
8DNM
 
 | | Human Brain Dihydropyrimidinase-related protein 2 | | Descriptor: | Dihydropyrimidinase-related protein 2 | | Authors: | Tringides, M.L. | | Deposit date: | 2022-07-11 | | Release date: | 2022-11-16 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | A cryo-electron microscopic approach to elucidate protein structures from human brain microsomes.
Life Sci Alliance, 6, 2023
|
|
7E3U
 
 | | Crystal structure of the Pseudomonas aeruginosa dihydropyrimidinase complexed with 5-AU | | Descriptor: | 5-AMINO-1H-PYRIMIDINE-2,4-DIONE, D-hydantoinase/dihydropyrimidinase, ZINC ION | | Authors: | Yang, Y.C, Luo, R.H, Huang, Y.H, Huang, C.Y, Lin, E.S. | | Deposit date: | 2021-02-09 | | Release date: | 2022-02-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.159 Å) | | Cite: | Molecular Insights into How the Dimetal Center in Dihydropyrimidinase Can Bind the Thymine Antagonist 5-Aminouracil: A Different Binding Mode from the Anticancer Drug 5-Fluorouracil.
Bioinorg Chem Appl, 2022, 2022
|
|
5UQC
 
 | | Crystal structure of mouse CRMP2 | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, Dihydropyrimidinase-related protein 2 | | Authors: | Khanna, M, Khanna, R, Perez-Miller, S, Francois-Moutal, L. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | A single structurally conserved SUMOylation site in CRMP2 controls NaV1.7 function.
Channels (Austin), 11, 2017
|
|
5HMD
 
 | |
4GBD
 
 | | Crystal Structure Of Adenosine Deaminase From Pseudomonas Aeruginosa Pao1 with bound Zn and methylthio-coformycin | | Descriptor: | (8R)-3-(5-S-methyl-5-thio-beta-D-ribofuranosyl)-3,6,7,8-tetrahydroimidazo[4,5-d][1,3]diazepin-8-ol, PHOSPHATE ION, Putative uncharacterized protein, ... | | Authors: | Ho, M, Guan, R, Almo, S.C, Schramm, V.L. | | Deposit date: | 2012-07-27 | | Release date: | 2013-06-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.975 Å) | | Cite: | Methylthioadenosine deaminase in an alternative quorum sensing pathway in Pseudomonas aeruginosa.
Biochemistry, 51, 2012
|
|
4UB9
 
 | | Structural and catalytic characterization of molinate hydrolase | | Descriptor: | Molinate hydrolase, ZINC ION | | Authors: | Leite, J.P, Duarte, M, Paiva, A, Ferreira-da-Silva, F, Matias, P.M, Nunes, O, Gales, L. | | Deposit date: | 2014-08-12 | | Release date: | 2015-06-24 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structure-guided engineering of molinate hydrolase for the degradation of thiocarbamate pesticides.
Plos One, 10, 2015
|
|
8JE0
 
 | | A novel amidohydrolase | | Descriptor: | 1,2-ETHANEDIOL, Amidase, ZINC ION | | Authors: | Ma, D, Feng, R. | | Deposit date: | 2023-05-15 | | Release date: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A novel amidohydrolase catalyze the degradation of PAM by Klebsiella sp. PCX
To Be Published
|
|
8YAG
 
 | | Cryo-electron microscopic structure of an amide hydrolase from Pseudoxanthomonas wuyuanensis | | Descriptor: | Imidazolonepropionase, ZINC ION | | Authors: | Dai, L.H, Xu, Y.H, Hu, Y.M, Niu, D, Yang, X.C, Shen, P.P, Li, X, Xie, Z.Z, Li, H, Guo, R.-T, Chen, C.-C. | | Deposit date: | 2024-02-09 | | Release date: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.33 Å) | | Cite: | Functional characterization and structural basis of an efficient ochratoxin A-degrading amidohydrolase.
Int.J.Biol.Macromol., 278, 2024
|
|
7UOF
 
 | | Dihydroorotase from M. jannaschii | | Descriptor: | Dihydroorotase, ZINC ION | | Authors: | Vitali, J, Nix, J.C, Newman, H.E, Colaneri, M.J. | | Deposit date: | 2022-04-12 | | Release date: | 2022-08-31 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of Methanococcus jannaschii dihydroorotase.
Proteins, 91, 2023
|
|
8WQ9
 
 | |
8J85
 
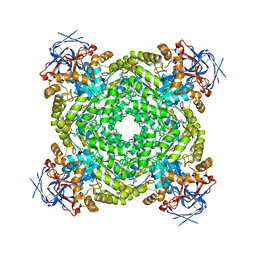 | | Cryo-EM structure of ochratoxin A-detoxifying amidohydrolase ADH3 mutant S88E in complex with ochratoxin A | | Descriptor: | (2~{S})-2-[[(3~{R})-5-chloranyl-3-methyl-8-oxidanyl-1-oxidanylidene-3,4-dihydroisochromen-7-yl]carbonylamino]-3-phenyl-propanoic acid, Amidohydrolase family protein, ZINC ION | | Authors: | Dai, L.H, Niu, D, Huang, J.-W, Li, X, Shen, P.P, Li, H, Hu, Y.M, Yang, Y, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2023-04-30 | | Release date: | 2023-08-30 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Cryo-EM structure and rational engineering of a superefficient ochratoxin A-detoxifying amidohydrolase.
J Hazard Mater, 458, 2023
|
|
7U5K
 
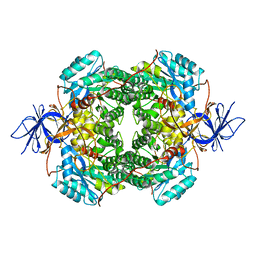 | | Cryo-EM Structure of DPYSL2 | | Descriptor: | Dihydropyrimidinase-related protein 2 | | Authors: | Morgan, C.E, Yu, E.W. | | Deposit date: | 2022-03-02 | | Release date: | 2022-12-14 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.78 Å) | | Cite: | Toward structural-omics of the bovine retinal pigment epithelium.
Cell Rep, 41, 2022
|
|
5HME
 
 | |
1J79
 
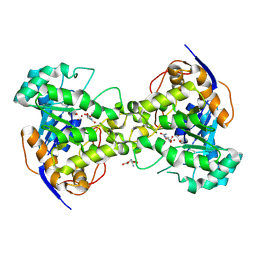 | | Molecular Structure of Dihydroorotase: A Paradigm for Catalysis Through the Use of a Binuclear Metal Center | | Descriptor: | N-CARBAMOYL-L-ASPARTATE, OROTIC ACID, ZINC ION, ... | | Authors: | Thoden, J.B, Phillips Jr, G.N, Neal, T.M, Raushel, F.M, Holden, H.M. | | Deposit date: | 2001-05-16 | | Release date: | 2001-06-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular structure of dihydroorotase: a paradigm for catalysis through the use of a binuclear metal center.
Biochemistry, 40, 2001
|
|
6JKU
 
 | | Crystal structure of N-acetylglucosamine-6-phosphate deacetylase from Pasteurella Multocida | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, N-acetylglucosamine-6-phosphate deacetylase, ... | | Authors: | Manjunath, L, Bose, S, Subramanian, R. | | Deposit date: | 2019-03-01 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Quaternary variations in the structural assembly of N-acetylglucosamine-6-phosphate deacetylase from Pasteurella multocida.
Proteins, 2020
|
|
1J6P
 
 | |
6JVB
 
 | |
4V1X
 
 | | The structure of the hexameric atrazine chlorohydrolase, AtzA | | Descriptor: | ATRAZINE CHLOROHYDROLASE, DI(HYDROXYETHYL)ETHER, FE (III) ION | | Authors: | Peat, T.S, Newman, J, Balotra, S, Lucent, D, Warden, A.C, Scott, C. | | Deposit date: | 2014-10-04 | | Release date: | 2015-03-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Structure of the Hexameric Atrazine Chlorohydrolase Atza.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
8IHS
 
 | | Cryo-EM structure of ochratoxin A-detoxifying amidohydrolase ADH3 in complex with ochratoxin A | | Descriptor: | (2~{S})-2-[[(3~{R})-5-chloranyl-3-methyl-8-oxidanyl-1-oxidanylidene-3,4-dihydroisochromen-7-yl]carbonylamino]-3-phenyl-propanoic acid, Amidohydrolase family protein, ZINC ION | | Authors: | Dai, L.H, Niu, D, Huang, J.-W, Li, X, Shen, P.P, Li, H, Hu, Y.M, Yang, Y, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2023-02-23 | | Release date: | 2023-08-30 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Cryo-EM structure and rational engineering of a superefficient ochratoxin A-detoxifying amidohydrolase.
J Hazard Mater, 458, 2023
|
|
8IHR
 
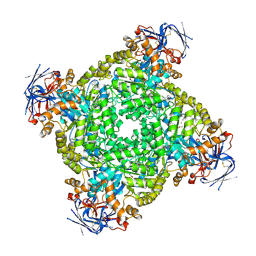 | | Cryo-EM structure of ochratoxin A-detoxifying amidohydrolase ADH3 in complex with Phe | | Descriptor: | Amidohydrolase family protein, PHENYLALANINE, ZINC ION | | Authors: | Dai, L.H, Niu, D, Huang, J.-W, Li, X, Shen, P.P, Li, H, Hu, Y.M, Yang, Y, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2023-02-23 | | Release date: | 2023-08-30 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Cryo-EM structure and rational engineering of a superefficient ochratoxin A-detoxifying amidohydrolase.
J Hazard Mater, 458, 2023
|
|
