6C1P
 
 | | HypoPP mutant | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CHAPSO, Ion transport protein, ... | | Authors: | Catterall, W.A, Zheng, N, Jiang, D, Gamal El-Din, T.M. | | Deposit date: | 2018-01-05 | | Release date: | 2018-05-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for gating pore current in periodic paralysis.
Nature, 557, 2018
|
|
8TG3
 
 | | tRNA 2'-phosphotransferase (Tpt1) from Aeropyrum pernix in complex with ADP-ribose-1" -phosphate | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, NITRATE ION, ... | | Authors: | Jacewicz, A, Dantuluri, S, Shuman, S. | | Deposit date: | 2023-07-12 | | Release date: | 2023-11-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structural basis for Tpt1-catalyzed 2'-PO 4 transfer from RNA and NADP(H) to NAD.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
6VGI
 
 | | Crystal Structures of FLAP bound to MK-866 | | Descriptor: | 3-[3-(tert-butylsulfanyl)-1-[(4-chlorophenyl)methyl]-5-(propan-2-yl)-1H-indol-2-yl]-2,2-dimethylpropanoic acid, 5-lipoxygenase-activating protein, SULFATE ION | | Authors: | Ho, J.D, Lee, M.R, Rauch, C.T, Aznavour, K, Park, J.S, Luz, J.G, Antonysamy, S, Condon, B, Maletic, M, Zhang, A, Hickey, M.J, Hughes, N.E, Chandrasekhar, S, Sloan, A.V, Gooding, K, Harvey, A, Yu, X.P, Kahl, S.D, Norman, B.H. | | Deposit date: | 2020-01-08 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structure-based, multi-targeted drug discovery approach to eicosanoid inhibition: Dual inhibitors of mPGES-1 and 5-lipoxygenase activating protein (FLAP).
Biochim Biophys Acta Gen Subj, 1865, 2020
|
|
6G25
 
 | | X-ray structure of NSD3-PWWP1 in complex with compound 4 | | Descriptor: | 3,5-dimethyl-4-(4-pyridin-4-yl-1~{H}-pyrazol-3-yl)-1,2-oxazole, Histone-lysine N-methyltransferase NSD3 | | Authors: | Boettcher, J, Muellauer, B.J, Weiss-Puxbaum, A, Zoephel, A. | | Deposit date: | 2018-03-22 | | Release date: | 2019-06-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.432 Å) | | Cite: | Fragment-based discovery of a chemical probe for the PWWP1 domain of NSD3.
Nat.Chem.Biol., 15, 2019
|
|
5EM2
 
 | | Crystal structure of the Erb1-Ytm1 complex | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Ribosome biogenesis protein ERB1, ... | | Authors: | Ahmed, Y.L, Sinning, I. | | Deposit date: | 2015-11-05 | | Release date: | 2015-12-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Concerted removal of the Erb1-Ytm1 complex in ribosome biogenesis relies on an elaborate interface.
Nucleic Acids Res., 44, 2016
|
|
8T1Q
 
 | | Crystal structure of human CPSF73 catalytic segment in complex with compound 1 | | Descriptor: | 3-[7,7-bis(oxidanyl)-8-oxa-7-boranuidabicyclo[4.3.0]nona-1,3,5-trien-5-yl]-~{N}-[3-(4-ethanoylphenyl)phenyl]propanamide, CHLORIDE ION, Cleavage and polyadenylation specificity factor subunit 3, ... | | Authors: | Huang, J, Tong, L. | | Deposit date: | 2023-06-02 | | Release date: | 2023-11-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Anticancer benzoxaboroles block pre-mRNA processing by directly inhibiting CPSF3.
Cell Chem Biol, 31, 2024
|
|
6HVD
 
 | | Human SLK bound to a maleimide inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 3-(1,3-benzothiazol-2-ylamino)-4-(2-methoxyphenyl)pyrrole-2,5-dione, STE20-like serine/threonine-protein kinase, ... | | Authors: | Sorrell, F.J, Berger, B.T, Salah, E, Serafim, R.A.M, Savitsky, P.A, Krojer, T, Bailey, H.J, Pinkas, D, Burgess-Brown, N.A, von Delft, F, Knapp, S, Arrowsmith, C, Bountra, C, Edwards, A.M, Elkins, J.M. | | Deposit date: | 2018-10-10 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Human SLK bound to a maleimide inhibitor
To Be Published
|
|
8JAM
 
 | |
6GA4
 
 | | Bacteriorhodopsin, 1 ps state, real-space refined against 15% extrapolated map | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, DECANE, ... | | Authors: | Nass Kovacs, G, Colletier, J.-P, Gruenbein, M.L, Stensitzki, T, Batyuk, A, Carbajo, S, Doak, R.B, Ehrenberg, D, Foucar, L, Gasper, R, Gorel, A, Hilpert, M, Kloos, M, Koglin, J, Reinstein, J, Roome, C.M, Schlesinger, R, Seaberg, M, Shoeman, R.L, Stricker, M, Boutet, S, Haacke, S, Heberle, J, Domratcheva, T, Schlichting, I. | | Deposit date: | 2018-04-11 | | Release date: | 2019-04-24 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Three-dimensional view of ultrafast dynamics in photoexcited bacteriorhodopsin.
Nat Commun, 10, 2019
|
|
6XKG
 
 | | Crystal structure of 3-O-Sulfotransferase isoform 3 in complex with 8mer oligosaccharide with 6S sulfation | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-6-O-sulfo-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid, 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid, ... | | Authors: | Pedersen, L.C, Liu, J, Wander, R. | | Deposit date: | 2020-06-26 | | Release date: | 2021-06-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Deciphering the substrate recognition mechanisms of the heparan sulfate 3- O -sulfotransferase-3.
Rsc Chem Biol, 2, 2021
|
|
5LYY
 
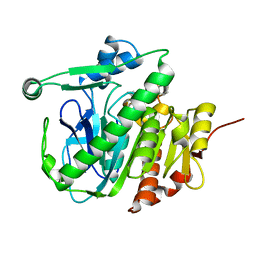 | | Fragment-based inhibitors of Lipoprotein associated Phospholipase A2 | | Descriptor: | 3-[2-(4-fluoranylphenoxy)ethyl]-1,3-diazaspiro[4.5]decane-2,4-dione, Platelet-activating factor acetylhydrolase | | Authors: | Woolford, A, Day, P. | | Deposit date: | 2016-09-29 | | Release date: | 2016-12-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Fragment-Based Approach to the Development of an Orally Bioavailable Lactam Inhibitor of Lipoprotein-Associated Phospholipase A2 (Lp-PLA2).
J. Med. Chem., 59, 2016
|
|
9GL7
 
 | | EGFR Exon20 insertion mutant NPG bound with (S)-3-((3-chloro-2-methoxyphenyl)amino)-2-(3-((tetrahydrofuran-2-yl)methoxy)pyridin-4-yl)-1,5,6,7-tetrahydro-4H-pyrrolo[3,2-c]pyridin-4-one | | Descriptor: | 3-[(3-chloranyl-2-methoxy-phenyl)amino]-2-[3-[[(2S)-oxolan-2-yl]methoxy]pyridin-4-yl]-1,5,6,7-tetrahydropyrrolo[3,2-c]pyridin-4-one, Epidermal growth factor receptor | | Authors: | Hilbert, B.J, Brooijmans, N, Milgram, B.C, Pagliarini, R.A. | | Deposit date: | 2024-08-27 | | Release date: | 2025-05-14 | | Last modified: | 2025-07-23 | | Method: | X-RAY DIFFRACTION (1.878 Å) | | Cite: | STX-721, a Covalent EGFR/HER2 Exon 20 Inhibitor, Utilizes Exon 20-Mutant Dynamic Protein States and Achieves Unique Mutant Selectivity Across Human Cancer Models.
Clin.Cancer Res., 31, 2025
|
|
6HDK
 
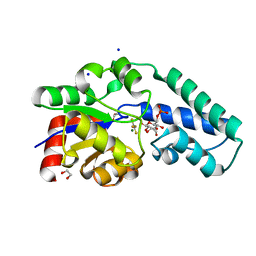 | |
6C7Y
 
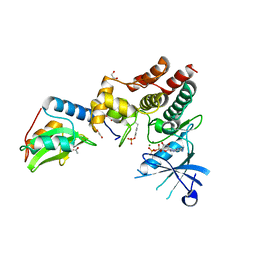 | | Crystal structure of inhibitory protein SOCS1 in complex with JAK1 kinase domain | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Liau, N.P.D, Laktyushin, A, Lucet, I.S, Murphy, J.M, Yao, S, Callaghan, K, Nicola, N.A, Kershaw, N.J, Babon, J.J. | | Deposit date: | 2018-01-23 | | Release date: | 2018-05-02 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.499 Å) | | Cite: | The molecular basis of JAK/STAT inhibition by SOCS1.
Nat Commun, 9, 2018
|
|
7CWG
 
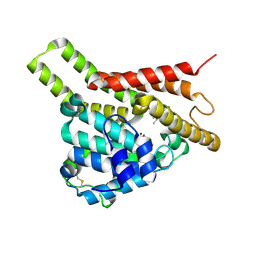 | | Crystal structure of PDE8A catalytic domain in complex with 3a | | Descriptor: | 9-[(1~{S})-1-[4-[2,2-bis(fluoranyl)ethoxy]pyridin-2-yl]ethyl]-2-chloranyl-purin-6-amine, High affinity cAMP-specific and IBMX-insensitive 3',5'-cyclic phosphodiesterase 8A, MAGNESIUM ION, ... | | Authors: | Huang, Y, Wu, X.-N, Zhou, Q, Wu, Y, Luo, H.-B. | | Deposit date: | 2020-08-28 | | Release date: | 2021-09-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Rational Design of 2-Chloroadenine Derivatives as Highly Selective Phosphodiesterase 8A Inhibitors.
J.Med.Chem., 63, 2020
|
|
8OIK
 
 | | D-PHAT domain (NTD) of human SAMD4A | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Protein Smaug homolog 1 | | Authors: | Kubikova, J, Jeske, M. | | Deposit date: | 2023-03-23 | | Release date: | 2023-04-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structural basis for binding of Drosophila Smaug to the GPCR Smoothened and to the germline inducer Oskar.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
6G3M
 
 | | Phosphotriesterase PTE_C23M_4 | | Descriptor: | 1-ethyl-1-methyl-cyclohexane, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Dym, O, Aggarwal, N, Albeck, S, Unger, T, Hamer Rogotner, S, Silman, I, Leader, H, Ashani, Y, Goldsmith, M, Greisen, P, Tawfik, D, Sussman, L.J. | | Deposit date: | 2018-03-26 | | Release date: | 2019-04-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.665 Å) | | Cite: | Phosphotriesterase
PTE_C23M_4
To Be Published
|
|
5ISG
 
 | | Crystal structure of mouse CARM1 in complex with inhibitor SA0707 | | Descriptor: | 1,2-DIMETHOXYETHANE, 1,2-ETHANEDIOL, 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, ... | | Authors: | Cura, V, Marechal, N, Mailliot, J, Troffer-Charlier, N, Hassenboehler, P, Wurtz, J.M, Bonnefond, L, Cavarelli, J. | | Deposit date: | 2016-03-15 | | Release date: | 2017-03-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.418 Å) | | Cite: | Crystal structure of mouse CARM1 in complex with inhibitor SA0707
To Be Published
|
|
6GBJ
 
 | | Repertoires of functionally diverse enzymes through computational design at epistatic active-site positions | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, Parathion hydrolase, ... | | Authors: | Khersonsky, O, Lipsh, R, Avizemer, Z, Goldsmith, M, Ashani, Y, Leader, H, Dym, O, Rogotner, S, Trudeau, D, Tawfik, D.S, Fleishman, S.J. | | Deposit date: | 2018-04-15 | | Release date: | 2018-10-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Automated Design of Efficient and Functionally Diverse Enzyme Repertoires.
Mol. Cell, 72, 2018
|
|
5DMY
 
 | | Beta-galactosidase - construct 33-930 | | Descriptor: | 1,2-ETHANEDIOL, Beta-galactosidase, MAGNESIUM ION, ... | | Authors: | Watson, K.A, Lazidou, A. | | Deposit date: | 2015-09-09 | | Release date: | 2016-10-05 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Rational protein engineering toward the development of a beta-galactosidase with improved functional properties
To Be Published
|
|
6ZSA
 
 | | Human mitochondrial ribosome bound to mRNA, A-site tRNA and P-site tRNA | | Descriptor: | 12S mitochondrial rRNA, 16S mitochondrial rRNA, 28S ribosomal protein S10, ... | | Authors: | Aibara, S, Singh, V, Modelska, A, Amunts, A. | | Deposit date: | 2020-07-15 | | Release date: | 2020-10-21 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural basis of mitochondrial translation.
Elife, 9, 2020
|
|
4Z4J
 
 | |
4X6M
 
 | |
6SIJ
 
 | | SAD structure of Hen Egg White Lysozyme recovered by continuous rotation data collection and multivariate analysis of Friedel pairs | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Garcia-Bonete, M.J, Katona, G. | | Deposit date: | 2019-08-10 | | Release date: | 2019-11-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.61018026 Å) | | Cite: | Bayesian machine learning improves single-wavelength anomalous diffraction phasing.
Acta Crystallogr.,Sect.A, 75, 2019
|
|
6UM9
 
 | |
