2JB8
 
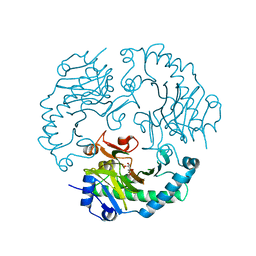 | | Deacetoxycephalosporin C synthase complexed with 5-hydroxy-4-keto valeric acid | | Descriptor: | 4,5-DIOXOPENTANOIC ACID, DEACETOXYCEPHALOSPORIN C SYNTHETASE, FE (III) ION | | Authors: | Cicero, G, Carbonera, C, Valegard, K, Hajdu, J, Andersson, I, Ranghino, G. | | Deposit date: | 2006-12-05 | | Release date: | 2008-02-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Study of the Oxidative Half-Reaction Catalysed by a Non-Heme Ferrous Catalytic Centre by Means of Structural and Computational Methodologies
Int.J.Quantum Chem., 107, 2007
|
|
2JHK
 
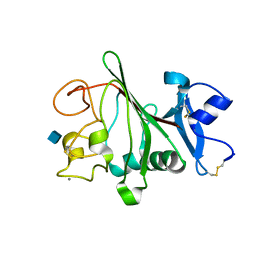 | | Structure of globular heads of M-ficolin complexed with N-acetyl-D- glucosamine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, FICOLIN-1 | | Authors: | Garlatti, V, Martin, L, Gout, E, Reiser, J.B, Arlaud, G.J, Thielens, N.M, Gaboriaud, C. | | Deposit date: | 2007-02-22 | | Release date: | 2007-10-09 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Basis for Innate Immune Sensing by M-Ficolin and its Control by a Ph-Dependent Conformational Switch.
J.Biol.Chem., 282, 2007
|
|
2JHI
 
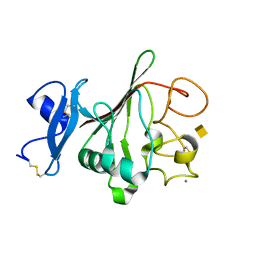 | | Structure of globular heads of M-ficolin complexed with N-acetyl-D- galactosamine | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, CALCIUM ION, FICOLIN-1 | | Authors: | Garlatti, V, Martin, L, Gout, E, Reiser, J.B, Arlaud, G.J, Thielens, N.M, Gaboriaud, C. | | Deposit date: | 2007-02-22 | | Release date: | 2007-10-09 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis for Innate Immune Sensing by M-Ficolin and its Control by a Ph-Dependent Conformational Switch.
J.Biol.Chem., 282, 2007
|
|
1HB9
 
 | | quasi-atomic resolution model of bacteriophage PRD1 wild type virion, obtained by combined cryo-EM and X-ray crystallography. | | Descriptor: | BACTERIOPHAGE PRD1 | | Authors: | San Martin, C, Burnett, R.M, De Haas, F, Heinkel, R, Rutten, T, Fuller, S.D, Butcher, S.J, Bamford, D.H. | | Deposit date: | 2001-04-13 | | Release date: | 2001-12-05 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (25 Å) | | Cite: | Combined Em/X-Ray Imaging Yields a Quasi-Atomic Model of the Adenovirus-Related Bacteriophage Prd1 and Shows Key Capsid and Membrane Interactions
Structure, 9, 2001
|
|
2ZBG
 
 | | Calcium pump crystal structure with bound AlF4 and TG in the absence of calcium | | Descriptor: | MAGNESIUM ION, OCTANOIC ACID [3S-[3ALPHA, 3ABETA, ... | | Authors: | Toyoshima, C, Ogawa, H, Norimatsu, Y. | | Deposit date: | 2007-10-20 | | Release date: | 2007-12-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | How processing of aspartylphosphate is coupled to lumenal gating of the ion pathway in the calcium pump
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
4N0E
 
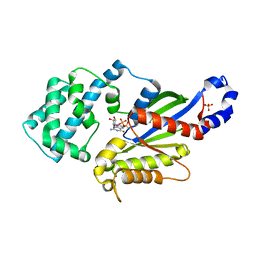 | | Crystal structure of the K345L variant of the Gi alpha1 subunit bound to GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Guanine nucleotide-binding protein G(i) subunit alpha-1, SULFATE ION | | Authors: | Thaker, T.M, Preininger, A.M, Sarwar, M, Hamm, H.E, Iverson, T.M. | | Deposit date: | 2013-10-01 | | Release date: | 2014-03-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Transient Interaction between the Phosphate Binding Loop and Switch I Contributes to the Allosteric Network between Receptor and Nucleotide in G alpha i1.
J.Biol.Chem., 289, 2014
|
|
3C6O
 
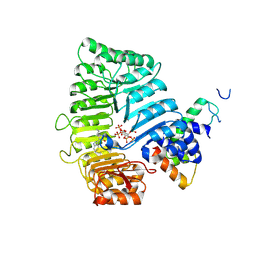 | |
2KDD
 
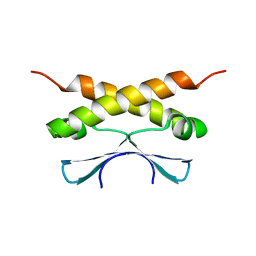 | |
2KBU
 
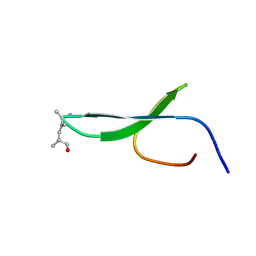 | |
2KEG
 
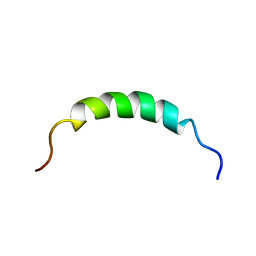 | | NMR structure of Plantaricin K in DPC-micelles | | Descriptor: | PlnK | | Authors: | Rogne, P, Haugen, M, Fimland, G, Nissen-Meyer, J, Kristiansen, P. | | Deposit date: | 2009-01-30 | | Release date: | 2009-07-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of the two-peptide bacteriocin plantaricin JK.
Peptides, 30, 2009
|
|
2KJX
 
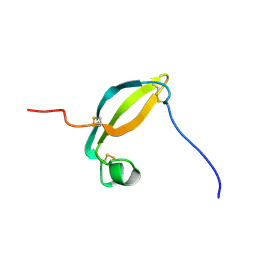 | | Solution structure of the extracellular domain of JTB | | Descriptor: | Jumping translocation breakpoint protein | | Authors: | Rousseau, F, Lingel, A, Pan, B, Fairbrother, W.J, Bazan, F. | | Deposit date: | 2009-06-10 | | Release date: | 2010-08-11 | | Last modified: | 2012-04-18 | | Method: | SOLUTION NMR | | Cite: | The structure of the extracellular domain of the jumping translocation breakpoint protein reveals a variation of the midkine fold.
J.Mol.Biol., 415, 2012
|
|
2D4C
 
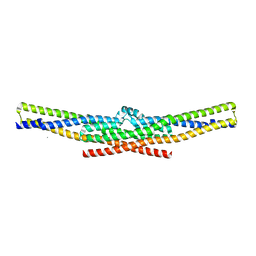 | | Crystal structure of the endophilin BAR domain mutant | | Descriptor: | CALCIUM ION, SH3-containing GRB2-like protein 2 | | Authors: | Masuda, M, Takeda, S. | | Deposit date: | 2005-10-13 | | Release date: | 2006-07-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Endophilin BAR domain drives membrane curvature by two newly identified structure-based mechanisms
Embo J., 25, 2006
|
|
2KLU
 
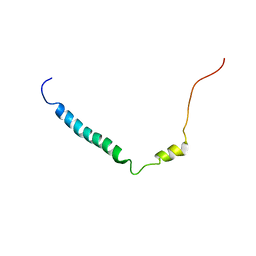 | |
3AFA
 
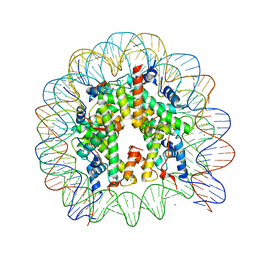 | | The human nucleosome structure | | Descriptor: | 146-MER DNA, CHLORIDE ION, Histone H2A type 1-B/E, ... | | Authors: | Tachiwana, H, Kagawa, W, Osakabe, A, Koichiro, K, Shiga, T, Kimura, H, Kurumizaka, H. | | Deposit date: | 2010-02-24 | | Release date: | 2010-05-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of instability of the nucleosome containing a testis-specific histone variant, human H3T
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
4QHL
 
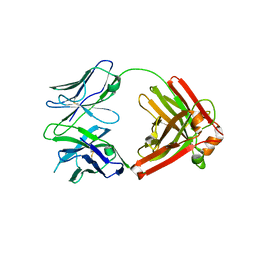 | | I3.2 (unbound) from CH103 Lineage | | Descriptor: | I3 heavy chain, UCA light chain | | Authors: | Fera, D, Harrison, S.C. | | Deposit date: | 2014-05-28 | | Release date: | 2014-06-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.153 Å) | | Cite: | Affinity maturation in an HIV broadly neutralizing B-cell lineage through reorientation of variable domains.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3A1D
 
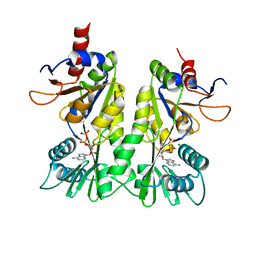 | | Crystal structure of the P- and N-domains of CopA, a copper-transporting P-type ATPase, bound with ADP-Mg | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Probable copper-exporting P-type ATPase A | | Authors: | Tsuda, T, Toyoshima, C. | | Deposit date: | 2009-03-31 | | Release date: | 2009-07-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Nucleotide recognition by CopA, a Cu+-transporting P-type ATPase.
Embo J., 28, 2009
|
|
3AI5
 
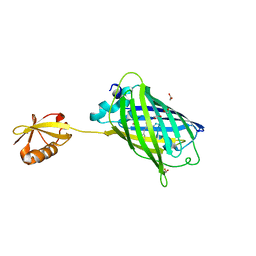 | | Crystal structure of yeast enhanced green fluorescent protein-ubiquitin fusion protein | | Descriptor: | 1,2-ETHANEDIOL, yeast enhanced green fluorescent protein,Ubiquitin | | Authors: | Suzuki, N, Wakatsuki, S, Kawasaki, M. | | Deposit date: | 2010-05-10 | | Release date: | 2010-09-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystallization of small proteins assisted by green fluorescent protein
Acta Crystallogr.,Sect.D, 66, 2010
|
|
1F3H
 
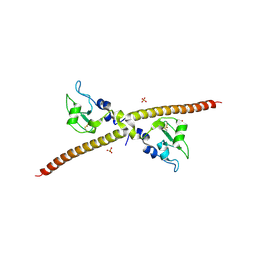 | | X-RAY CRYSTAL STRUCTURE OF THE HUMAN ANTI-APOPTOTIC PROTEIN SURVIVIN | | Descriptor: | SULFATE ION, SURVIVIN, ZINC ION | | Authors: | Verdecia, M.A, Huang, H, Dutil, E, Hunter, T, Noel, J.P. | | Deposit date: | 2000-06-03 | | Release date: | 2000-12-06 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Structure of the human anti-apoptotic protein survivin reveals a dimeric arrangement.
Nat.Struct.Biol., 7, 2000
|
|
3C8X
 
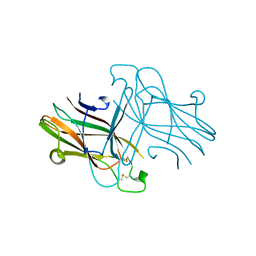 | | Crystal structure of the ligand binding domain of human Ephrin A2 (Epha2) receptor protein kinase | | Descriptor: | Ephrin type-A receptor 2 | | Authors: | Walker, J.R, Yermekbayeva, L, Seitova, A, Butler-Cole, C, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-02-14 | | Release date: | 2008-03-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Architecture of Eph receptor clusters.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2FLD
 
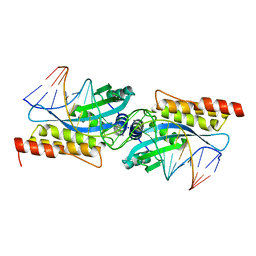 | | I-MsoI Re-Designed for Altered DNA Cleavage Specificity | | Descriptor: | 5'-D(*CP*GP*GP*AP*AP*CP*GP*GP*TP*CP*TP*CP*AP*CP*GP*AP*CP*CP*TP*TP*CP*TP*GP*C)-3', 5'-D(*GP*CP*AP*GP*AP*AP*GP*GP*TP*CP*GP*TP*GP*AP*GP*AP*CP*CP*GP*TP*TP*CP*CP*G)-3', CALCIUM ION, ... | | Authors: | Ashworth, J, Duarte, C.M, Havranek, J.J, Sussman, D, Monnat, R.J, Stoddard, B.L, Baker, D. | | Deposit date: | 2006-01-05 | | Release date: | 2006-06-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Computational redesign of endonuclease DNA binding and cleavage specificity.
Nature, 441, 2006
|
|
2GK7
 
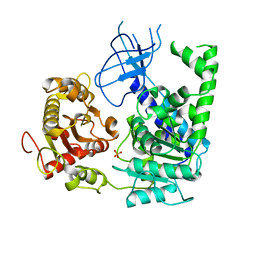 | | Structural and Functional insights into the human Upf1 helicase core | | Descriptor: | PHOSPHATE ION, Regulator of nonsense transcripts 1 | | Authors: | Cheng, Z, Muhlrad, D, Parker, R, Song, H. | | Deposit date: | 2006-03-31 | | Release date: | 2007-01-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and functional insights into the human Upf1 helicase core
Embo J., 26, 2007
|
|
3C6G
 
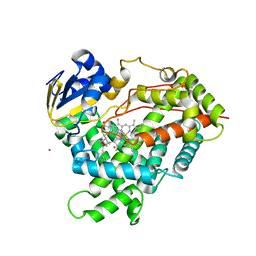 | | Crystal structure of CYP2R1 in complex with vitamin D3 | | Descriptor: | (1S,3Z)-3-[(2E)-2-[(1R,3AR,7AS)-7A-METHYL-1-[(2R)-6-METHYLHEPTAN-2-YL]-2,3,3A,5,6,7-HEXAHYDRO-1H-INDEN-4-YLIDENE]ETHYLI DENE]-4-METHYLIDENE-CYCLOHEXAN-1-OL, Cycloheptakis-(1-4)-(alpha-D-glucopyranose), Cytochrome P450 2R1, ... | | Authors: | Strushkevich, N.V, Min, J, Loppnau, P, Tempel, W, Arrowsmith, C.H, Edwards, A.M, Sundstrom, M, Weigelt, J, Bochkarev, A, Plotnikov, A.N, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-02-04 | | Release date: | 2008-02-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural analysis of CYP2R1 in complex with vitamin D3.
J.Mol.Biol., 380, 2008
|
|
2GK6
 
 | | Structural and Functional insights into the human Upf1 helicase core | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Cheng, Z, Muhlrad, D, Parker, R, Song, H. | | Deposit date: | 2006-03-31 | | Release date: | 2007-01-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and functional insights into the human Upf1 helicase core
Embo J., 26, 2007
|
|
3BND
 
 | | Lipoxygenase-1 (Soybean), I553V Mutant | | Descriptor: | FE (III) ION, Seed lipoxygenase-1 | | Authors: | Tomchick, D.R. | | Deposit date: | 2007-12-14 | | Release date: | 2008-04-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Enzyme structure and dynamics affect hydrogen tunneling: the impact of a remote side chain (I553) in soybean lipoxygenase-1.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2DEF
 
 | |
