4PRJ
 
 | |
2ZJY
 
 | | Structure of the K349P mutant of Gi alpha 1 subunit bound to ALF4 and GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Guanine nucleotide-binding protein G(i), alpha-1 subunit, ... | | Authors: | Morikawa, T, Muroya, A, Sugio, S, Wakamatsu, K, Kohno, T. | | Deposit date: | 2008-03-11 | | Release date: | 2009-03-24 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | How GPCRs activate G proteins: Structural changes form C-terminal tail to GDP binding pocket
To be Published
|
|
4PSH
 
 | | Structure of holo ArgBP from T. maritima | | Descriptor: | ABC-type transporter, periplasmic subunit family 3, ARGININE | | Authors: | Ruggiero, A, Dattelbaum, J.D, Staiano, M, Berisio, R, D'Auria, S, Vitagliano, L. | | Deposit date: | 2014-03-07 | | Release date: | 2014-07-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A loose domain swapping organization confers a remarkable stability to the dimeric structure of the arginine binding protein from Thermotoga maritima
Plos One, 9, 2014
|
|
2KLM
 
 | | Solution Structure of L11 with SAXS and RDC | | Descriptor: | 50S ribosomal protein L11 | | Authors: | Wang, J, Zuo, X, Yu, P, Schwieters, C.D, Wang, Y. | | Deposit date: | 2009-07-06 | | Release date: | 2009-10-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | Determination of multicomponent protein structures in solution using global orientation and shape restraints.
J.Am.Chem.Soc., 131, 2009
|
|
2KHO
 
 | |
3B5Z
 
 | | Crystal Structure of MsbA from Salmonella typhimurium with ADP Vanadate | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Lipid A export ATP-binding/permease protein msbA, VANADATE ION | | Authors: | Ward, A, Reyes, C.L, Yu, J, Roth, C.B, Chang, G. | | Deposit date: | 2007-10-26 | | Release date: | 2007-12-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Flexibility in the ABC transporter MsbA: Alternating access with a twist.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2K6A
 
 | | Solution structure of EAS D15 truncation mutant | | Descriptor: | Hydrophobin | | Authors: | Kwan, A.H. | | Deposit date: | 2008-07-07 | | Release date: | 2008-08-19 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | The Cys3-Cys4 loop of the hydrophobin EAS is not required for rodlet formation and surface activity.
J.Mol.Biol., 382, 2008
|
|
1G5Q
 
 | | EPID H67N COMPLEXED WITH SUBSTRATE PEPTIDE DSYTC | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, EPIDERMIN MODIFYING ENZYME EPID, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Blaesse, M, Kupke, T, Huber, R, Steinbacher, S. | | Deposit date: | 2000-11-02 | | Release date: | 2001-05-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Crystal structure of the peptidyl-cysteine decarboxylase EpiD complexed with a pentapeptide substrate.
EMBO J., 19, 2000
|
|
2B0Z
 
 | |
3BI1
 
 | |
2I5N
 
 | | 1.96 A X-ray structure of photosynthetic reaction center from Rhodopseudomonas viridis:Crystals grown by microfluidic technique | | Descriptor: | 15-cis-1,2-dihydroneurosporene, BACTERIOCHLOROPHYLL B, BACTERIOPHEOPHYTIN B, ... | | Authors: | Li, L, Mustafi, D, Fu, Q, Tereshko, V, Chen, D.L, Tice, J.D, Ismagilov, R.F. | | Deposit date: | 2006-08-25 | | Release date: | 2006-09-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Nanoliter microfluidic hybrid method for simultaneous screening and optimization validated with crystallization of membrane proteins.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2I4B
 
 | | Crystal structure of Bicarbonate Transport Protein CmpA from Synechocystis sp. PCC 6803 in complex with bicarbonate and calcium | | Descriptor: | BICARBONATE ION, Bicarbonate transporter, CALCIUM ION | | Authors: | Koropatkin, N.M, Smith, T.J, Pakrasi, H.B. | | Deposit date: | 2006-08-21 | | Release date: | 2006-12-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | The Structure of a Cyanobacterial Bicarbonate Transport Protein, CmpA.
J.Biol.Chem., 282, 2007
|
|
1GNC
 
 | |
2IBF
 
 | |
4PRS
 
 | | Structure of apo ArgBP from T. maritima | | Descriptor: | ABC-type transporter, periplasmic subunit family 3 | | Authors: | Ruggiero, A, Dattelbaum, J.D, Staiano, M, Berisio, R, D'Auria, S, Vitagliano, L. | | Deposit date: | 2014-03-06 | | Release date: | 2014-07-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | A loose domain swapping organization confers a remarkable stability to the dimeric structure of the arginine binding protein from Thermotoga maritima
Plos One, 9, 2014
|
|
2IAG
 
 | | Crystal structure of human prostacyclin synthase | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Prostacyclin synthase, SODIUM ION | | Authors: | Chiang, C.-W, Yeh, H.-C, Wang, L.-H, Chan, N.-L. | | Deposit date: | 2006-09-08 | | Release date: | 2006-10-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal Structure of the Human Prostacyclin Synthase
J.Mol.Biol., 364, 2006
|
|
2I49
 
 | |
2I8C
 
 | | Allosteric inhibition of Staphylococcus aureus D-alanine:D-alanine ligase revealed by crystallographic studies | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, D-alanine-D-alanine ligase, MAGNESIUM ION, ... | | Authors: | Liu, S, Chang, J.S, Herberg, J.T, Horng, M, Tomich, P.K, Lin, A.H, Marotti, K.R. | | Deposit date: | 2006-09-01 | | Release date: | 2006-09-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Allosteric inhibition of Staphylococcus aureus D-alanine:D-alanine ligase revealed by crystallographic studies.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
1HB7
 
 | | quasi-atomic resolution model of bacteriophage PRD1 sus1 mutant, obtained by combined cryo-EM and X-ray crystallography. | | Descriptor: | BACTERIOPHAGE PRD1 SUS1 MUTANT CAPSID | | Authors: | San Martin, C, Burnett, R.M, De Haas, F, Heinkel, R, Rutten, T, Fuller, S.D, Butcher, S.J, Bamford, D.H. | | Deposit date: | 2001-04-12 | | Release date: | 2001-12-05 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (14 Å) | | Cite: | Combined Em/X-Ray Imaging Yields a Quasi-Atomic Model of the Adenovirus-Related Bacteriophage Prd1 and Shows Key Capsid and Membrane Interactions.
Structure, 9, 2001
|
|
2I48
 
 | |
2HXH
 
 | | KIF1A head-microtubule complex structure in adp-form | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Kikkawa, M, Hirokawa, N. | | Deposit date: | 2006-08-03 | | Release date: | 2006-10-10 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (11 Å) | | Cite: | High-resolution cryo-EM maps show the nucleotide binding pocket of KIF1A in open and closed conformations
Embo J., 25, 2006
|
|
1H76
 
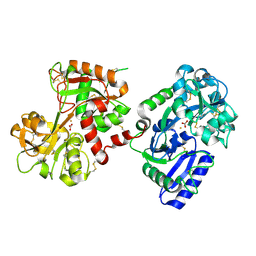 | | The crystal structure of diferric porcine serum transferrin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CARBONATE ION, FE (III) ION, ... | | Authors: | Hall, D.R, Hadden, J.M, Leonard, G.A, Bailey, S, Neu, M, Winn, M, Lindley, P.F. | | Deposit date: | 2001-07-03 | | Release date: | 2002-01-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The Crystal and Molecular Structures of Diferric Porcine and Rabbit Serum Transferrins at Resolutions of 2.15 And 2.60A, Respectively
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1GU6
 
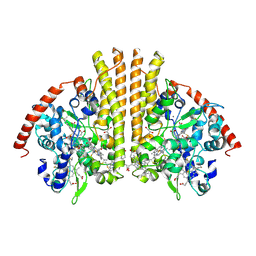 | | Structure of the Periplasmic Cytochrome c Nitrite Reductase from Escherichia coli | | Descriptor: | CALCIUM ION, CYTOCHROME C552, GLYCEROL, ... | | Authors: | Bamford, V.A, Angove, H.C, Seward, H.E, Thomson, A.J, Cole, J.A, Butt, J.N, Hemmings, A.M, Richardson, D.J. | | Deposit date: | 2002-01-24 | | Release date: | 2002-03-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure and Spectroscopy of the Periplasmic Cytochrome C Nitrite Reductase from Escherichia Coli
Biochemistry, 41, 2002
|
|
3B5W
 
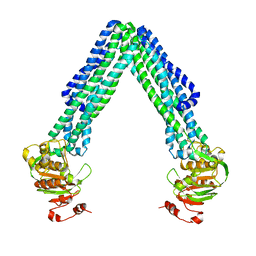 | | Crystal Structure of Eschericia coli MsbA | | Descriptor: | Lipid A export ATP-binding/permease protein msbA | | Authors: | Ward, A, Reyes, C.L, Yu, J, Roth, C.B, Chang, G. | | Deposit date: | 2007-10-26 | | Release date: | 2007-12-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (5.3 Å) | | Cite: | Flexibility in the ABC transporter MsbA: Alternating access with a twist.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2GJF
 
 | |
