8F4W
 
 | | Crystal structure of acetyltransferase Eis from M. tuberculosis in complex with venlafaxine | | Descriptor: | 1-[(1R)-2-(dimethylamino)-1-(4-methoxyphenyl)ethyl]cyclohexan-1-ol, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | Authors: | Pang, A.H, Punetha, A, Garneau-Tsodikova, S, Tsodikov, O.V. | | Deposit date: | 2022-11-11 | | Release date: | 2023-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery and Mechanistic Analysis of Structurally Diverse Inhibitors of Acetyltransferase Eis among FDA-Approved Drugs.
Biochemistry, 62, 2023
|
|
8OG8
 
 | | Crystal structure of human DCAF1 WD40 repeats (Q1250L) in complex with compound 3 | | Descriptor: | 1,2-ETHANEDIOL, 5-(2-methyl-1-phenyl-propan-2-yl)imidazo[2,1-a]isoquinoline, ACETATE ION, ... | | Authors: | Schroeder, M, Vulpetti, A, Renatus, M. | | Deposit date: | 2023-03-19 | | Release date: | 2023-06-14 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Discovery of New Binders for DCAF1, an Emerging Ligase Target in the Targeted Protein Degradation Field.
Acs Med.Chem.Lett., 14, 2023
|
|
9BV6
 
 | | Crystal structure of human CYP3A4 in complex with SJ000388260 | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, 5-(4-methoxyphenyl)-1-[4-(2-methyl-1,3-thiazol-4-yl)phenyl]-1H-1,2,3-triazole, Cytochrome P450 3A4, ... | | Authors: | Jingheng, W, Nithianantham, S, Miller, D.J, Chen, T. | | Deposit date: | 2024-05-20 | | Release date: | 2025-04-16 | | Last modified: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Decoding the selective chemical modulation of CYP3A4.
Nat Commun, 16, 2025
|
|
5LTU
 
 | | Crystal Structure of NUDT4A- Diphosphoinositol polyphosphate phosphohydrolase 2 | | Descriptor: | 1,2-ETHANEDIOL, Diphosphoinositol polyphosphate phosphohydrolase 2 | | Authors: | Srikannathasan, V, Nunez, C.A, Tallant, C, Siejka, P, Mathea, S, Newman, J, Strain-Damerell, C, Elkins, J.M, Burgess-Brown, N, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F, Huber, K. | | Deposit date: | 2016-09-07 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Crystal Structure of Human NUDT4A- Diphosphoinositol polyphosphate phosphohydrolase 2
To Be Published
|
|
8OGB
 
 | | Crystal structure of human DCAF1 WD40 repeats (Q1250L) in complex with compound 8 | | Descriptor: | 1,2-ETHANEDIOL, DDB1- and CUL4-associated factor 1, DIMETHYL SULFOXIDE, ... | | Authors: | Schroeder, M, Vulpetti, A, Renatus, M. | | Deposit date: | 2023-03-19 | | Release date: | 2023-06-14 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Discovery of New Binders for DCAF1, an Emerging Ligase Target in the Targeted Protein Degradation Field.
Acs Med.Chem.Lett., 14, 2023
|
|
6S4R
 
 | | The crystal structure of glycogen phosphorylase in complex with 11 | | Descriptor: | (2~{R},3~{S},4~{R},5~{R},6~{S})-2-(hydroxymethyl)-6-(2-naphthalen-2-yl-1,3-thiazol-4-yl)oxane-3,4,5-triol, Glycogen phosphorylase, muscle form, ... | | Authors: | Kyriakis, E, Papaioannou, O.S.E, Solovou, T.G.A, Skamnaki, V.T, Leonidas, D.D. | | Deposit date: | 2019-06-28 | | Release date: | 2020-02-19 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The architecture of hydrogen and sulfur sigma-hole interactions explain differences in the inhibitory potency of C-beta-d-glucopyranosyl thiazoles, imidazoles and an N-beta-d glucopyranosyl tetrazole for human liver glycogen phosphorylase and offer new insights to structure-based design.
Bioorg.Med.Chem., 28, 2020
|
|
7A5I
 
 | | Structure of the human mitoribosome with A- P-and E-site mt-tRNAs | | Descriptor: | 12S rRNA, 16S rRNA, 28S ribosomal protein S10, ... | | Authors: | Desai, N, Yang, H, Chandrasekaran, V, Kazi, R, Minczuk, M, Ramakrishnan, V. | | Deposit date: | 2020-08-21 | | Release date: | 2020-12-23 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Elongational stalling activates mitoribosome-associated quality control.
Science, 370, 2020
|
|
5LU0
 
 | | Crystal structure of H. pylori referent strain in complex with PO4 | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, PHOSPHATE ION, ... | | Authors: | Stefanic, Z. | | Deposit date: | 2016-09-07 | | Release date: | 2017-09-20 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Helicobacter pylori purine nucleoside phosphorylase shows new distribution patterns of open and closed active site conformations and unusual biochemical features.
FEBS J., 285, 2018
|
|
8Y4I
 
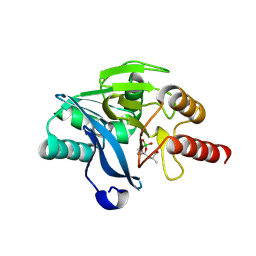 | | Metal Beta-Lactamase VIM-2 in Complex with MBL inhibitor B7 | | Descriptor: | 2-[[(9R)-2-fluoranyl-7,7-bis(oxidanyl)-8-oxa-7-boranuidabicyclo[4.3.0]nona-1,3,5-trien-9-yl]methyl]prop-2-enoic acid, FORMIC ACID, Metal Beta-Lactamase VIM-2, ... | | Authors: | Li, G.-B, Wang, S.-Y. | | Deposit date: | 2024-01-30 | | Release date: | 2025-02-05 | | Last modified: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (1.935 Å) | | Cite: | Discovery of 2-((1-hydroxy-1,3-dihydrobenzo[c][1,2]oxaborol-3-yl)methyl)acrylic acids as potent MBL inhibitors
To Be Published
|
|
6S72
 
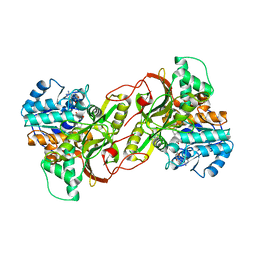 | |
6S70
 
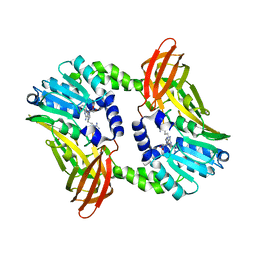 | | Crystal structure of CARM1 in complex with inhibitor UM251 | | Descriptor: | 1-[5-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl-(3-azanylpropyl)amino]pentyl]guanidine, GLYCEROL, Histone-arginine methyltransferase CARM1 | | Authors: | Gunnell, E.A, Muhsen, U, Dowden, J, Dreveny, I. | | Deposit date: | 2019-07-04 | | Release date: | 2020-03-04 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and biochemical evaluation of bisubstrate inhibitors of protein arginine N-methyltransferases PRMT1 and CARM1 (PRMT4).
Biochem.J., 477, 2020
|
|
5LGP
 
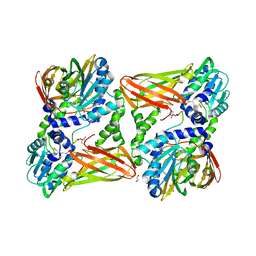 | | Crystal structure of mouse CARM1 in complex with ligand P1C3s | | Descriptor: | (2~{R},3~{R},4~{S},5~{R})-2-(6-aminopurin-9-yl)-5-propyl-oxolane-3,4-diol, 1,2-ETHANEDIOL, Histone-arginine methyltransferase CARM1, ... | | Authors: | Marechal, N, Troffer-Charlier, N, Cura, V, Bonnefond, L, Cavarelli, J. | | Deposit date: | 2016-07-08 | | Release date: | 2017-03-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Transition state mimics are valuable mechanistic probes for structural studies with the arginine methyltransferase CARM1.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
7OF4
 
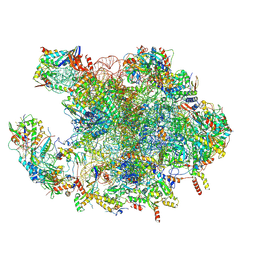 | | Structure of mature human mitochondrial ribosome large subunit in complex with GTPBP6 (PTC conformation 1). | | Descriptor: | 16S ribosomal RNA, 39S ribosomal protein L10, mitochondrial, ... | | Authors: | Hillen, H.S, Lavdovskaia, E, Nadler, F, Hanitsch, E, Linden, A, Bohnsack, K.E, Urlaub, H, Richter-Dennerlein, R. | | Deposit date: | 2021-05-04 | | Release date: | 2021-06-09 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural basis of GTPase-mediated mitochondrial ribosome biogenesis and recycling.
Nat Commun, 12, 2021
|
|
5CS6
 
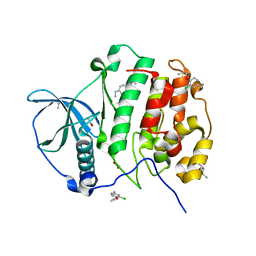 | | Crystal Structure of CK2alpha with Compound 3 bound | | Descriptor: | 1-(3-chloro-4-propoxyphenyl)methanamine, ACETATE ION, Casein kinase II subunit alpha, ... | | Authors: | Brear, P, De Fusco, C, Georgiou, K.H, Spring, D, Hyvonen, M. | | Deposit date: | 2015-07-23 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Specific inhibition of CK2 alpha from an anchor outside the active site.
Chem Sci, 7, 2016
|
|
8D11
 
 | |
5JDH
 
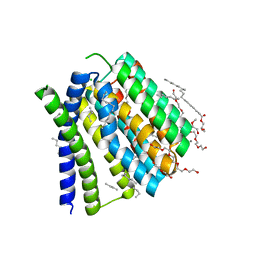 | | Structural mechanisms of extracellular ion exchange and induced binding-site occlusion in the sodium-calcium exchanger NCX_Mj soaked with 10 mM Na+ and 10mM Ca2+ | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, CALCIUM ION, ... | | Authors: | Liao, J, Jiang, Y.X, Faraldo-Gomez, J.D. | | Deposit date: | 2016-04-16 | | Release date: | 2016-05-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.203 Å) | | Cite: | Mechanism of extracellular ion exchange and binding-site occlusion in a sodium/calcium exchanger.
Nat.Struct.Mol.Biol., 23, 2016
|
|
7QG5
 
 | | IRAK4 in complex with inhibitor | | Descriptor: | 4-[(1-methylcyclopropyl)amino]-2-[[1-(1-methylpiperidin-4-yl)pyrazol-4-yl]amino]-6-pyrimidin-5-yl-pyrido[4,3-d]pyrimidin-5-one, Interleukin-1 receptor-associated kinase 4 | | Authors: | Xue, Y, Aagaard, A, Robb, G.R, Degorce, S.L. | | Deposit date: | 2021-12-07 | | Release date: | 2022-05-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification and optimisation of a pyrimidopyridone series of IRAK4 inhibitors.
Bioorg.Med.Chem., 63, 2022
|
|
6HJK
 
 | | Crystal Structure of Aurora-A L210C catalytic domain in complex with ASDO2 | | Descriptor: | (~{E})-~{N}-[4-(4-azanyl-1-propan-2-yl-pyrazolo[3,4-d]pyrimidin-3-yl)phenyl]-4-[4-fluoranyl-3-(trifluoromethyl)phenyl]-4-oxidanylidene-but-2-enamide, Aurora kinase A, CHLORIDE ION, ... | | Authors: | Bayliss, R, McIntyre, P.J. | | Deposit date: | 2018-09-04 | | Release date: | 2018-10-03 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Type II Kinase Inhibitors Targeting Cys-Gatekeeper Kinases Display Orthogonality with Wild Type and Ala/Gly-Gatekeeper Kinases.
ACS Chem. Biol., 13, 2018
|
|
1FHB
 
 | | THREE-DIMENSIONAL SOLUTION STRUCTURE OF THE CYANIDE ADDUCT OF A MET80ALA VARIANT OF SACCHAROMYCES CEREVISIAE ISO-1-CYTOCHROME C. IDENTIFICATION OF LIGAND-RESIDUE INTERACTIONS IN THE DISTAL HEME CAVITY | | Descriptor: | CYANIDE ION, FERRICYTOCHROME C, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Banci, L, Bertini, I, Bren, K.L, Gray, H.B, Sompornpisut, P, Turano, P. | | Deposit date: | 1995-06-16 | | Release date: | 1995-09-15 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Three-Dimensional Solution Structure of the Cyanide Adduct of a met80Ala Variant of Saccharomyces Cerevisiae Iso-1-Cytochrome C. Identification of Ligand-Residue Interactions in the Distal Heme Cavity
Biochemistry, 34, 1995
|
|
7VH6
 
 | | Cryo-EM structure of the hexameric plasma membrane H+-ATPase in the active state (pH 6.0, BeF3-, conformation 1, C1 symmetry) | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, BERYLLIUM TRIFLUORIDE ION, Plasma membrane ATPase 1 | | Authors: | Zhao, P, Zhao, C, Chen, D, Yun, C, Li, H, Bai, L. | | Deposit date: | 2021-09-21 | | Release date: | 2021-11-24 | | Last modified: | 2025-09-17 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure and activation mechanism of the hexameric plasma membrane H + -ATPase.
Nat Commun, 12, 2021
|
|
7AJA
 
 | | Structure of DYRK1A in complex with compound 28 | | Descriptor: | 1-[4-(1-benzofuran-5-yl)pyridin-2-yl]piperazine, CHLORIDE ION, Dual specificity tyrosine-phosphorylation-regulated kinase 1A | | Authors: | Dokurno, P, Surgenor, A.E, Kotschy, A. | | Deposit date: | 2020-09-28 | | Release date: | 2021-05-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Guided Discovery of Potent and Selective DYRK1A Inhibitors.
J.Med.Chem., 64, 2021
|
|
8OEO
 
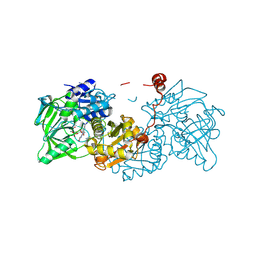 | | Aspergillus niger ferulic acid decarboxylase (Fdc) V186C-A296C (DB4) variant in complex with prenylated flavin | | Descriptor: | 1-deoxy-5-O-phosphono-1-(3,3,4,5-tetramethyl-9,11-dioxo-2,3,8,9,10,11-hexahydro-7H-quinolino[1,8-fg]pteridin-12-ium-7-y l)-D-ribitol, Ferulic acid decarboxylase 1, MANGANESE (II) ION, ... | | Authors: | Roberts, G.W, Leys, D. | | Deposit date: | 2023-03-10 | | Release date: | 2023-09-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Aspergillus niger ferulic acid decarboxylase (Fdc) V186C-A296C (DB4) variant in complex with prenylated flavin
To Be Published
|
|
8OEH
 
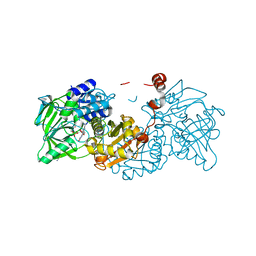 | | Aspergillus niger ferulic acid decarboxylase (Fdc) C122-S261C (DB3) variant in complex with prenylated flavin | | Descriptor: | 1-deoxy-5-O-phosphono-1-(3,3,4,5-tetramethyl-9,11-dioxo-2,3,8,9,10,11-hexahydro-7H-quinolino[1,8-fg]pteridin-12-ium-7-y l)-D-ribitol, Ferulic acid decarboxylase 1, MANGANESE (II) ION, ... | | Authors: | Roberts, G.W, Leys, D. | | Deposit date: | 2023-03-10 | | Release date: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Aspergillus niger ferulic acid decarboxylase (Fdc) C122-S261C (DB3) variant in complex with prenylated flavin
To Be Published
|
|
8YKY
 
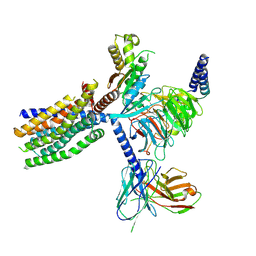 | | Structure of human class T GPCR TAS2R14-Ggustducin complex with agonist 28.1 | | Descriptor: | 4-methyl-N-[(2M)-2-(1H-tetrazol-5-yl)phenyl]-6-(trifluoromethyl)pyrimidin-2-amine, CHOLESTEROL, G alpha gustducin protein, ... | | Authors: | Hu, X.L, Wu, L.J, Hua, T, Liu, Z.J. | | Deposit date: | 2024-03-05 | | Release date: | 2024-07-10 | | Last modified: | 2025-07-16 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Bitter taste TAS2R14 activation by intracellular tastants and cholesterol.
Nature, 631, 2024
|
|
7MLC
 
 | | PYL10 bound to the ABA pan-antagonist 4a | | Descriptor: | 1-{2-[3,5-dicyclopropyl-4-(4-{[(quinoxaline-2-carbonyl)amino]methyl}-1H-1,2,3-triazol-1-yl)phenyl]acetamido}cyclohexane-1-carboxylic acid, Abscisic acid receptor PYL10, GLYCEROL | | Authors: | Peterson, F.C, Vaidya, A.S, Volkman, B.F, Cutler, S.R. | | Deposit date: | 2021-04-28 | | Release date: | 2021-09-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Click-to-lead design of a picomolar ABA receptor antagonist with potent activity in vivo.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
