1ED3
 
 | | CRYSTAL STRUCTURE OF RAT MINOR HISTOCOMPATIBILITY ANTIGEN COMPLEX RT1-AA/MTF-E. | | Descriptor: | BETA-2-MICROGLOBULIN, CLASS I MAJOR HISTOCOMPATIBILITY ANTIGEN RT1-AA, PEPTIDE MTF-E (13N3E) | | Authors: | Speir, J.A, Stevens, J, Joly, E, Butcher, G.W, Wilson, I.A. | | Deposit date: | 2000-01-26 | | Release date: | 2001-02-28 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Two different, highly exposed, bulged structures for an unusually long peptide bound to rat MHC class I RT1-Aa.
Immunity, 14, 2001
|
|
3MA7
 
 | | Crystal structure of Cardiolipin bound to mouse CD1D | | Descriptor: | (2R,5R,11R,14R)-5,8,11-trihydroxy-5,11-dioxido-17-oxo-2,14-bis(tetradecanoyloxy)-4,6,10,12,16-pentaoxa-5,11-diphosphatriacont-1-yl tetradecanoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zajonc, D.M. | | Deposit date: | 2010-03-23 | | Release date: | 2011-03-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Cardiolipin Binds to CD1d and Stimulates CD1d-Restricted {gamma}{delta} T Cells in the Normal Murine Repertoire.
J.Immunol., 186, 2011
|
|
1EC5
 
 | | CRYSTAL STRUCTURE OF FOUR-HELIX BUNDLE MODEL | | Descriptor: | PROTEIN (FOUR-HELIX BUNDLE MODEL), ZINC ION | | Authors: | Geremia, S. | | Deposit date: | 2000-01-25 | | Release date: | 2000-07-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Inaugural article: retrostructural analysis of metalloproteins: application to the design of a minimal model for diiron proteins.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
3NWM
 
 | | Crystal structure of a single chain construct composed of MHC class I H-2Kd, beta-2microglobulin and a peptide which is an autoantigen for type 1 diabetes | | Descriptor: | Peptide/beta-2microglobulin/MHC class I H-2Kd chimeric protein | | Authors: | Ramagopal, U.A, Samanta, D, Nathenson, S.G, Almo, S.C. | | Deposit date: | 2010-07-09 | | Release date: | 2011-07-06 | | Last modified: | 2017-06-28 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and functional characterization of a single-chain peptide-MHC molecule that modulates both naive and activated CD8+ T cells.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3O8X
 
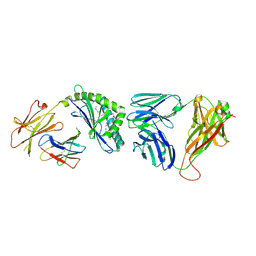 | | Recognition of Glycolipid Antigen by iNKT Cell TCR | | Descriptor: | (2S,3R)-3-HYDROXY-2-(TETRADECANOYLAMINO)OCTADECYL ALPHA-D-GALACTOPYRANOSIDURONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zajonc, D.M, Li, Y. | | Deposit date: | 2010-08-03 | | Release date: | 2010-09-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | The V alpha 14 invariant natural killer T cell TCR forces microbial glycolipids and CD1d into a conserved binding mode.
J.Exp.Med., 207, 2010
|
|
3O9W
 
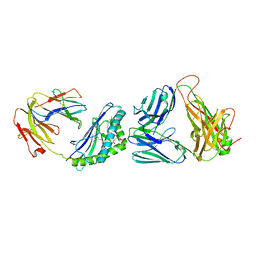 | | Recognition of a Glycolipid Antigen by the iNKT Cell TCR | | Descriptor: | (2S)-3-(alpha-D-galactopyranosyloxy)-2-(hexadecanoyloxy)propyl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zajonc, D.M, Li, Y. | | Deposit date: | 2010-08-04 | | Release date: | 2010-09-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The V alpha 14 invariant natural killer T cell TCR forces microbial glycolipids and CD1d into a conserved binding mode.
J.Exp.Med., 207, 2010
|
|
8JHW
 
 | |
3P4N
 
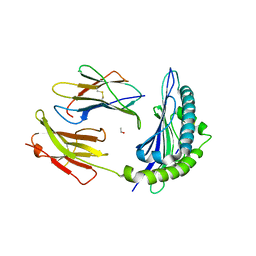 | | Crystal Structure of H2-Kb in complex with the NP205-PV epitope YTVKFPNM, an 8-mer peptide from PV | | Descriptor: | ACETYL GROUP, Beta-2-microglobulin, H-2 class I histocompatibility antigen, ... | | Authors: | Gras, S, Guillonneau, C, Rossjohn, J. | | Deposit date: | 2010-10-06 | | Release date: | 2011-10-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of H2-Kb in complex with the NP205-PV epitope YTVKFPNM, an 8-mer peptide from PV
To be Published
|
|
3P4M
 
 | | Crystal Structure of H2-Kb in complex with the NP205-LCMV epitope YTVKYPNL, an 8-mer peptide from the LCMV | | Descriptor: | ACETYL GROUP, Beta-2-microglobulin, H-2 class I histocompatibility antigen, ... | | Authors: | Gras, S, Guillonneau, C, Rossjohn, J. | | Deposit date: | 2010-10-06 | | Release date: | 2011-10-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of H2-Kb in complex with the NP205-LCMV epitope YTVKYPNL, an 8-mer peptide from the LCMV
To be Published
|
|
8JHV
 
 | |
3PAB
 
 | | Crystal Structure of H2-Kb in complex with a mutant of the chicken ovalbumin epitope OVA-E1 | | Descriptor: | Beta-2-microglobulin, H-2 class I histocompatibility antigen, K-B alpha chain, ... | | Authors: | Wesselingh, R, Gras, S, Guillonneau, C, Turner, S.J, Rossjohn, J. | | Deposit date: | 2010-10-19 | | Release date: | 2011-10-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Affinity thresholds for naive CD8+ CTL activation by peptides and engineered influenza A viruses
J.Immunol., 187, 2011
|
|
3P9M
 
 | | Crystal Structure of H2-Kb in complex with a mutant of the chicken ovalbumin epitope OVA-G4 | | Descriptor: | Beta-2-microglobulin, H-2 class I histocompatibility antigen, K-B alpha chain, ... | | Authors: | Wesselingh, R, Gras, S, Guillonneau, C, Turner, S.J, Rossjohn, J. | | Deposit date: | 2010-10-18 | | Release date: | 2011-10-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Affinity thresholds for naive CD8+ CTL activation by peptides and engineered influenza A viruses
J.Immunol., 187, 2011
|
|
3P4O
 
 | | Crystal Structure of H2-Kb in complex with the mutant NP205-LCMV-V3A epitope YTAKYPNL, an 8-mer modified peptide from the LCMV | | Descriptor: | ACETYL GROUP, Beta-2-microglobulin, H-2 class I histocompatibility antigen, ... | | Authors: | Gras, S, Guillonneau, C, Rossjohn, J. | | Deposit date: | 2010-10-06 | | Release date: | 2011-10-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of H2-Kb in complex with the mutant NP205-LCMV-V3A epitope YTAKYPNL, an 8-mer modified peptide from the LCMV
To be Published
|
|
3P9L
 
 | | Crystal Structure of H2-Kb in complex with the chicken ovalbumin epitope OVA | | Descriptor: | Beta-2-microglobulin, CALCIUM ION, H-2 class I histocompatibility antigen, ... | | Authors: | Wesselingh, R, Gras, S, Guillonneau, C, Turner, S.J, Rossjohn, J. | | Deposit date: | 2010-10-18 | | Release date: | 2011-10-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Affinity thresholds for naive CD8+ CTL activation by peptides and engineered influenza A viruses
J.Immunol., 187, 2011
|
|
2X50
 
 | | Crystal structure of Trypanothione reductase from Leishmania infantum in complex with NADPH and silver | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SILVER ION, ... | | Authors: | Baiocco, P, Ilari, A, Colotti, G. | | Deposit date: | 2010-02-03 | | Release date: | 2011-01-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Inhibitory Effect of Silver Nanoparticles on Trypanothione Reductase Activity and Leishmania Infantum Proliferation
Acs Med.Chem.Lett., 2, 2011
|
|
2F7N
 
 | | Structure of D. radiodurans Dps-1 | | Descriptor: | COBALT (II) ION, DNA-binding stress response protein, Dps family, ... | | Authors: | Lee, Y.H, Kim, S.G, Bhattacharyya, G, Grove, A. | | Deposit date: | 2005-12-01 | | Release date: | 2006-11-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of Dps-1, a functionally distinct Dps protein from Deinococcus radiodurans.
J.Mol.Biol., 361, 2006
|
|
3PWU
 
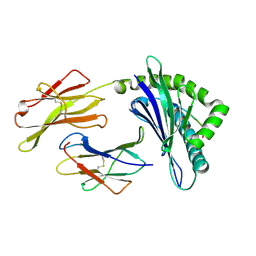 | | An immmunodominant CTL epitope from rinderpest virus presented by cattle MHC class I molecule N*01801(BoLA-A11) | | Descriptor: | Beta-2-microglobulin, IPA from Hemagglutinin glycoprotein, MHC class I antigen | | Authors: | Li, X, Liu, J, Qi, J, Gao, F, Li, Q, Li, X, Zhang, N, Xia, C, Gao, G.F. | | Deposit date: | 2010-12-09 | | Release date: | 2011-08-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.899 Å) | | Cite: | Two distinct conformations of a rinderpest virus epitope presented by bovine major histocompatibility complex class I N*01801: a host strategy to present featured peptides
J.Virol., 85, 2011
|
|
2W9R
 
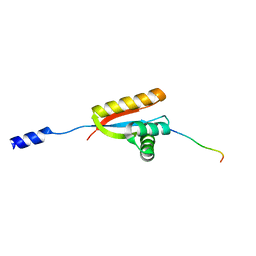 | | Structural basis of N-end rule substrate recognition in Escherichia coli by the ClpAP adaptor protein ClpS | | Descriptor: | ATP-DEPENDENT CLP PROTEASE ADAPTER PROTEIN CLPS, DNA PROTECTION DURING STARVATION PROTEIN | | Authors: | Schuenemann, V, Kralik, S.M, Albrecht, R, Spall, S.K, Truscott, K.N, Dougan, D.A, Zeth, K. | | Deposit date: | 2009-01-28 | | Release date: | 2009-04-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis of N-End Rule Substrate Recognition in Escherichia Coli by the Clpap Adaptor Protein Clps.
Embo Rep., 10, 2009
|
|
2YJK
 
 | |
2YJJ
 
 | |
1S7U
 
 | | Crystal structures of the murine class I major histocompatibility complex H-2Db in complex with LCMV-derived gp33 index peptide and three of its escape variants | | Descriptor: | Beta-2-microglobulin, Glycoprotein 9-residue peptide, H-2 class I histocompatibility antigen, ... | | Authors: | Velloso, L.M, Michaelsson, J, Ljunggren, H.G, Schneider, G, Achour, A. | | Deposit date: | 2004-01-30 | | Release date: | 2004-05-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Determination of structural principles underlying three different modes of lymphocytic choriomeningitis virus escape from CTL recognition.
J.Immunol., 172, 2004
|
|
4G43
 
 | | Structure of the chicken MHC class I molecule BF2*0401 complexed to P5E | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 8-MERIC PEPTIDE P5E, Beta-2 microglobulin, ... | | Authors: | Zhang, J, Chen, Y, Qi, J, Gao, F, Liu, J, Kaufman, J, Xia, C, Gao, G.F. | | Deposit date: | 2012-07-16 | | Release date: | 2012-11-21 | | Method: | X-RAY DIFFRACTION (1.803 Å) | | Cite: | Narrow Groove and Restricted Anchors of MHC Class I Molecule BF2*0401 Plus Peptide Transporter Restriction Can Explain Disease Susceptibility of B4 Chickens.
J.Immunol., 189, 2012
|
|
1NAM
 
 | | MURINE ALLOREACTIVE SCFV TCR-PEPTIDE-MHC CLASS I MOLECULE COMPLEX | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BM3.3 T Cell Receptor alpha-Chain, BM3.3 T Cell Receptor beta-Chain, ... | | Authors: | Reiser, J.-B, Darnault, C, Gregoire, C, Mosser, T, Mazza, G, Kearnay, A, van der Merwe, P.A, Fontecilla-Camps, J.C, Housset, D, Malissen, B. | | Deposit date: | 2002-11-28 | | Release date: | 2003-03-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | CDR3 loop flexibility contributes to the degeneracy of TCR recognition
Nat.Immunol., 4, 2003
|
|
5TJE
 
 | | Murine class I major histocompatibility complex H-2Db in complex with LCMV-derived gp33 and T cell receptor P14 | | Descriptor: | ALPHA CHAIN OF MURINE T CELL RECEPTOR p14, BETA CHAIN OF MURINE T CELL RECEPTOR p14, Beta-2-microglobulin, ... | | Authors: | Achour, A, Sandalova, T, Allerbring, E, Popov, A. | | Deposit date: | 2016-10-04 | | Release date: | 2017-11-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Thernary complexes of TCR P14 give insights into the mechanisms behind reestablishment of CTL responses against a viral escape mutant
to be published
|
|
5T7G
 
 | | Crystal Structure of Murine MHC-I H-2Dd in complex with Murine Beta2-Microglobulin and a Variant of Peptide (PT9) of HIV gp120 MN Isolate (IGPGRAFYT) | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, H-2 class I histocompatibility antigen, ... | | Authors: | Jiang, J, Natarajan, K, Margulies, D. | | Deposit date: | 2016-09-04 | | Release date: | 2017-10-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.961 Å) | | Cite: | Effects of Cross-Presentation, Antigen Processing, and Peptide Binding in HIV Evasion of T Cell Immunity.
J. Immunol., 200, 2018
|
|
